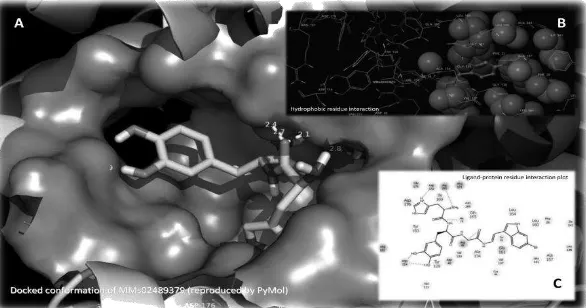
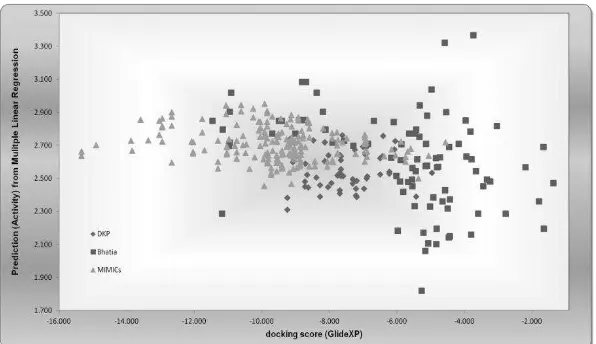
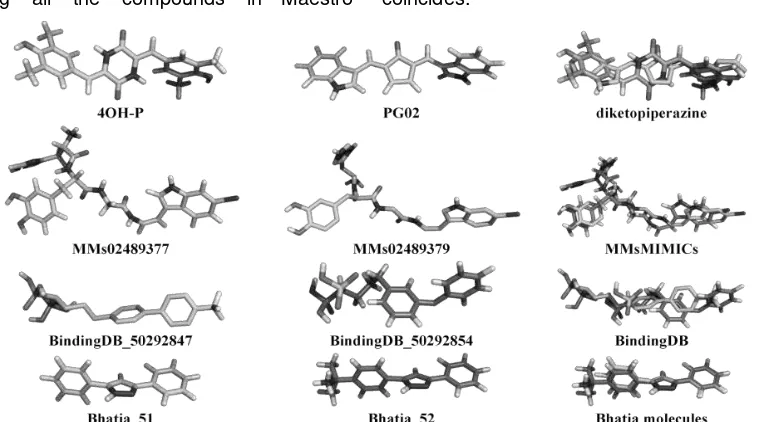
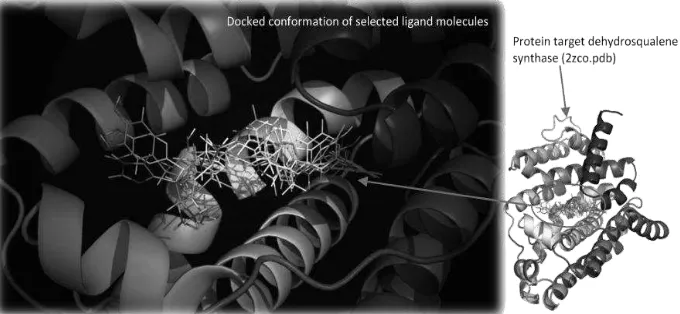
EXPLORING 3D MOLECULAR STUDIES OF DIKETOPIPERAZINE ANALOGUES ON Staphylococcus aureus DEHYDROSQUALENE SYNTHASE USING GLIDE-XP
Bebas
10
0
0
Teks penuh
Gambar


+3
Dokumen terkait