COMPARATIVE PHYLOGEOGRAPHY OF JAVA AND BALI
FRESHWATER FISHES USING DNA BARCODES
ARIEF ADITYA HUTAMA
GRADUATE SCHOOL
BOGOR AGRICULTURAL UNIVERSITY BOGOR
STATEMENT LETTER
I hereby declare that thesis entitled Comparative Phylogeography of Java and Bali Freshwater Fishes Using DNA Barcodes is based on original results of my own research supervised under advisory committee and has never been submitted in any form at any institution before. All information from other authors cited here are mentioned in the text and listed in the reference at the end part of the thesis.
Bogor, January 2016
SUMMARY
ARIEF ADITYA HUTAMA. Comparative Phylogeography of Java and Bali Freshwater Fishes Using DNA Barcodes. Supervised by BAMBANG SURYOBROTO, NICOLAS HUBERT, ACHMAD FARAJALLAH.
The information about the origin of the ichthyodiversity in Java is still poorly understood and spatio-temporal dynamic also still unknown. Therefore, biogeographic studies aiming at exploring the spatio-temporal dynamics of settlement of freshwater fishes in Java are certainly needed. Fishes are the most diverse group of vertebrates, and because of their large phenotypic diversity and profound changes during their ontogenetic development, fish identification is difficult.
DNA barcoding is a new approach to species identification based on a short fragment of DNA from a standardized region of the mitochondrial genome. In this study, we aim at using DNA barcoding to document and characterize the intra-specific diversity of several widespread species of freshwater fishes in Java and Bali. The objective is to explore the population structure of each species, identify common patterns and propose potential mechanisms to explain observed pattern. Due the high diversity of habitat in the study area, this research was conducted in a total of fifty one locations in Java and Bali islands in order to cover altitudinal and ecological gradients. Three species were selected as model: Channa gachua (Perciformes, Channidae), Glyptothorax platypogon( Siluriformes, Sisoridae), and Barbodes binotatus (Cypriniformes, Cyprinidae).
C. gachua, G. platypogon and B. binotatus display a high population genetic structure shown by a high genetic differentiation of the populations and the presence of several genetic clusters. High genetic variations among groups but low genetic variability within population according to nucleotidic diversity is likely to be the consequence of an important fragmentation of the populations as a high number of closely related haplotypes were observed within populations and genetic clusters. The age estimates of each genealogy suggest each species originated during the Pliocene at 3.05 Ma, 3 Ma and 2.6 Ma for C. gachua, G.platypogon and B. barbodes, respectively, the colonization of Java and Bali islands happened mainly during the Pleistocene. The lack of significance of the mantel tests showed that the differentiation of the populations was not the result of geographic distance but a consequence of habitat fragmentation. A genetic differentiation between the West and East populations was observed for all three species and a dynamic of secondary contact was observed in Central Java.
RINGKASAN
ARIEF ADITYA HUTAMA. Perbandingan Filogeografi dari Ikan Air Tawar di Jawa dan Bali Menggunakan Barcode DNA. Dibimbing oleh BAMBANG SURYOBROTO, NICOLAS HUBERT, ACHMAD FARAJALLAH.
Informasi mengenai asal – usul keragaman diversitas ikan serta dinamika keadaan geologi terkait waktu di pulau Jawa dan Bali masih kurang dipahami. Oleh karena itu, dibutuhkan studi tentang biogeografi yang bertujuan untuk mempelajari dinamika geologi terkait waktu pada kolonisasi ikan air tawar di pulau Jawa dan Bali Ikan merupakan grup vertebratra yang paling beragam, memiliki diversitas fenotipik yang tinggi dan adanya perubahan selama perkembangan ontogenetik, identifikasi ikan merupakan hal yang sulit.
DNA barcoding merupakan pendekatan baru untuk mengidentifikasi spesies didasarkan pada fragmen pendek DNA yang berasal dari bagian mitokondria yang telah dibakukan. Penelitian ini bertujuan menggunakan DNA barcoding untuk mendokumentasi dan mengkarakterisasi keragaman intraspesifik dari beberapa spesies ikan air tawar di pulau Jawa dan Bali yang memiliki wilayah persebaran luas. Sasaran utama dari penelitian ini adalah untuk mengetahui struktur populasi dari setiap spesies, mengidentifikasi pola dan mengajukan mekanisme potensial untuk menjelaskan pola yang teridentifikasi. Pengambilan sampel untuk penelitian dilaksanakan di 51 lokasi di pulau Jawa dan Bali dengan tujuan untuk mewakili diversitas habitat di pulau Jawa dan Bali yang tinggi. Tiga spesies dipilih menjadi model untuk penelitian ini : Channa gachua (Perciformes, Channidae), Glyptothorax platypogon ( Siluriformes, Sisoridae), and Barbodes binotatus (Cypriniformes, Cyprinidae).
C. gachua, G. platypogon and B. binotatus menunjukan tingginya struktur genetika populasi. Hal ini dapat dilihat dari tingginya diferensiasi populasi genetik dan keberadaan dari beberapa kluster genetic. Tingginya variasi genetik antar grup namun rendah di dalam satu populasi yang sama menurut diversitas nukleotida diperkirakan karena adanya fragmentasi populasi yang dapat dilihat dari tingginya nilai diversitas haplotipe namun jarak genetiknya kecil pada satu populasi yang sama. Perkiraan waktu secara genealogy memperkirakan ketiga spesies tersebut telah ada semenjak masa Pliocene yakni 3,05 juta tahun yang lalu untuk C. gachua, 3juta tahun yang lalu intuk G.platypogon dan 2,6 juta tahun yang lalu untuk B. barbodes dan berkolonisasi di pulau Jawa dan Bali pada masa Pleistocene. Hasil yang tidak signifikan dari mantel test menunjukkan bahwa diferensiasi populasi bukan disebabkan karena jarak geografi, namun dikarenakan adanya fragmentasi habitat. Adanya diferensiasi genetic antara populasi di Jawa Barat dan populasi di Jawa Timur terdapat pada ketiga spesies tersebut serta diperkirakan adanya secondary contact yang terjadi di daerah Jawa Tengah.
Copyright © 2016 Bogor Agricultural University
All Rights Reserved
It is prohibited to cite all or a part of this thesis without referring to and mentioning the source. Citation is permitted for the purposes of education, research, scientific paper, report, or critism writing only; and it does not defame the name and honour of
Bogor Agricultural University.
COMPARATIVE PHYLOGEOGRAPHY OF JAVA AND BALI
FRESHWATER FISHES USING DNA BARCODES
ARIEF ADITYA HUTAMA
Thesis
Submitted in partial fulfillment of the requirements for a Master Degree
in Animal Bioscience Major
Graduate School of Bogor Agricultural University
GRADUATE SCHOOL
BOGOR AGRICULTURAL UNIVERSITY BOGOR
ENDORSEMENT PAGE
Title : Comparative Phylogeography of Java and Bali Freshwater Fishes Using DNA Barcodes
Name : Arief Aditya Hutama NIM : G352130201
Major : Animal Bioscience
Endorsed by, Supervisor Committee
Dr. Bambang Suryobroto Chair
Dr. Nicolas Hubert Committee
Dr. Ir. Achmad Farajallah, M.Si Committee
Acknowledged by, Coordinator of Major
Animal Bioscience
Dr.Ir.R R Dyah Perwitasari, M.Sc.
p.p. Dean of Postgraduate School Secretary of Master Program
Prof. Dr.Ir. Nahrowi, M.Sc.
x
PREFACE
This research is a part of the general research program Inventory of Indonesian freshwater fishes through DNA Barcoding jointly developed by the Institut de Recherche pour le Développement (IRD, France) and the Indonesian Institute of Sciences (LIPI, Indonesia). This research program has been started in 2012 with the aim to characterize the diversity of Indonesian freshwater fishes through DNA barcoding. This international collaboration is ruled by a Memorandum of Understanding signed on 29th of October 2012 by the Chairman of LIPI (Prof. Dr. Likman Hakim) and the president of IRD (Prof. Dr. Michel Laurent). The research developed in the present thesis had been conducted from December 2012 to November 2015 in the context of this joint research program and as such, this research and dissemination of the results are bound by the obligations defined by the Memorandum of understanding between IRD and LIPI.
I want to send my gratitude to my supervisor Dr. Bambang Suryobroto, Dr. Nicolas Hubert and Dr. Achmad Farajallah for all the support. I wish thank to all of the IRD and LIPI staff that gave me the opportunity to develop this research: Dr. Renny Hadiaty, Sopian Sauri, Sumanta and Hadi Darhuddin. A gratitude to Mr. M Syamsul Arifin Zein, M.Si for allowing and helping me while I was working in Genetic Laboratory, Research Center for Biology LIPI, Cibinong. Thanks to all of the Animal Bioscience 2013, especially Esa and Maulana. Special thanks to my parents, my brother, and my sister for their love and support. I thank Allah SWT for all the blessings in my life.
Bogor, January 2016
xii
CONTENTS
SUMMARY ... iv
ENDORSEMENT PAGE ... ix
PREFACE ... x
CONTENTS ... xii
LIST OF TABLES ... xiii
LIST OF FIGURES ... xiii
LIST OF APPENDIX ... xiii
INTRODUCTION ... 1
MATERIAL AND METHOD ... 2
Geological Framework ... 2
Sample Collection ... 3
Genetic Analyses ... 4
Data Analyses ... 5
Phylogenetic Analyses of Gene genealogies ... 5
Population Structure Analyses ... 5
RESULT ... 6
Phylogenetic Analyses... 6
Population structure ... 11
DISCUSSION ... 13
CONCLUSION ... 15
REFERENCES ... 16
APPENDIX ... 18
xiii
LIST OF TABLES
1 Amova statistics and fixation indexes ... 11 2 Genetic diversity ... 13
LIST OF FIGURES
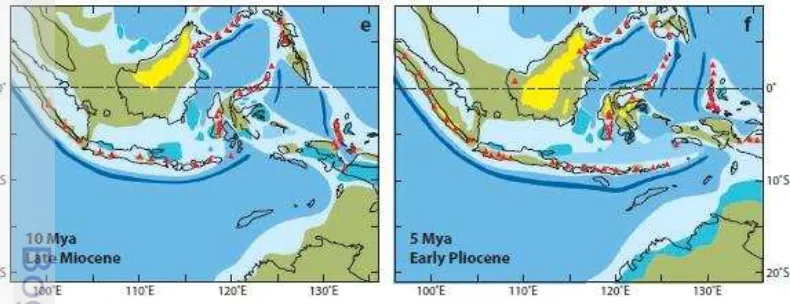
1 Late Miocene and Early Pliocene reconstructions of land and sea in the Indo-Australian Archipelago (Lohmann et al 2011) ... 2 2 Pleistocene map of South East Asia illustrating depth contours of 120m below
present sea levels (Voris 2000) ... 3 3 Sampling locations in Java and Bali islands. One point may represent several
sampling sites. ... 4 4 The three hierarchical levels of population structure and fixation index ... 6 5 Maximum Likehood tree of haplotypes for Channa gachua (5A), Glyptothorax
platypogon (5B) and Barbodes binotatus (5C). ... 9 6 Neighbour-joining tree of the population of C. gachua (6A), G. platypogon (6B)
and B. binotatus (6C). SAMOVA result of C. gachua (6D), G. platypogon (6E) and B. binotatus (6F) with CT(green), ST( red) and SC(blue), number of group as y-axis and variance number as x-axis... 10 7 Relationship between geographic distance and corrected pairwise Fst
(Fst(1-Fst)) in C. gachua (7A), G. platypogon (7B) and B. binotatus (7C) ... 12 8 Postulated distribution of land and sea in South East Asia from Hall (1998) ... 14
LIST OF APPENDIX
INTRODUCTION
Phylogeography is a field of research concerned with the principles and processes governing the geographic distributions of genetic diversity, either within or among closely related species. Phylogeography deals with the historical origin of the spatial distribution of genes. Analysing and interpreting the distribution of genetic diversity usually requires extensive input from population genetics, phylogeny, paleontology and geology. Thus, phylogeography is an integrative field of research that lies at an important crossroad among diverse microevolutionary and macroevolutionary disciplines (Avise 2001). Phylogeographic studies of freshwater fishes are still scarce in Indonesia. Thus, the present study may be expected to produce valuable insights into the biogeography of Indonesian aquatic biotas.
Among the 25 world‟s terrestrial hotspots, Indonesian hotspots (Sundaland, Wallacea) are currently the world‟s most threaten by human activities (Lamoureux et al 2006, Hoffman et al 2010). This is particularly evident in Java island where habitat destruction for palm oil plantation, species introduction through the ornamental fish market and pollution have deeply affected freshwater ecosystems. A previous study of extinction rates in West Java determined that 92.5 % and 76.5% of the fish species were extirpated, in the Ciliwung and Cisadane rivers, respectively (Hadiaty 2011). The origin of the ichthyodiversity in Java still poorly understood and spatio-temporal dynamics are also unknown. Therefore, biogeographic studies aiming at exploring the spatio-temporal dynamics of settlement of freshwater fishes in Java are certainly needed.
Given the diversity of ecological habitats in Java, cryptic diversity (two or more biological species previously assigned to the same taxon due to their morphological similarity) may be expected. The close morphological similarity among closely related and cryptic species makes them difficult to distinguish based on morphological characters and this issue currently limits the usefulness of a traditional taxonomy approach based on morphological characters. For instance, two new species from Java have been recently described based on the combination of morphological and molecular characters (Keith et al. 2014a, 2014b). Fishes are the most diverse group of vertebrates, and because of their large phenotypic diversity and profound changes during their ontogenetic development, fish identification is difficult.
2
Koizumi et al. 2012). The substitution rate in COI is high enough to allow the discrimination of not only closely related species, but also phylogeographic groups within a single species.
In this study, we aim at evaluating the utility of DNA barcoding to document and characterize the intra-specific diversity of several widespread species of freshwater fishes in Java and Bali. The objective is to explore the factors and mechanisms that contributed to population structure. To that end, the genetic diversity and population genetic structure in three widespread species has been used to estimate the impact of the geological on phylogeographic patterns and population structures.
MATERIAL AND METHOD
Geological Framework
Java and bali islands are part of Sundaland, which during the low sea levels had half the coastline and twice the land area as today. Sundaland has diverse and highly endemic biotas, although, it only consists of 4% of the planet‟s land area. At the beginning of the Cenozoic approximately 65 Mya, Sundaland was a continental promontory at the southern end of Eurasia (Hall 1996). Northward subduction of the Australian plate resumed beneath Indonesia, causing widespread volcanism at the active margin and producing chains of islands (arcs) similar to those of the West Pacific today. Sundaland was surrounded by subduction zones until the Miocene and was probably mainly close to sea level, with elevated areas in western Borneo and the Thai-Malay Peninsula (Lohmann et al 2011).
3
In the Early Miocene approximately 23 Mya, the Australian plate made contact with the submerged Sundaland margin near Sulawesi, and the Sunda region began to rotate, initially keeping pace with Australia‟s northward movement. There was a gradual increase in the area of shallow seas and reduction in land area on Sundaland until the Pliocene approximately 5 Mya). During the Quaternary, Australia continued to move north, and subduction-related deformation led to the gradual emergence of Sumatra and Java as major land areas during the Pliocene (Figure 1).
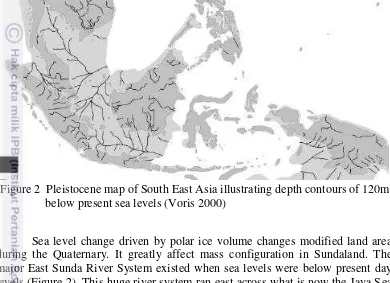
Figure 2 Pleistocene map of South East Asia illustrating depth contours of 120m below present sea levels (Voris 2000)
Sea level change driven by polar ice volume changes modified land area during the Quaternary. It greatly affect mass configuration in Sundaland. The major East Sunda River System existed when sea levels were below present day levels (Figure 2). This huge river system ran east across what is now the Java Sea to enter the sea near Bali. This system included virtually all the modern day river of the north coast of Java, south coast of Borneo and the northern portion of the east coast of Sumatra (Lohmann et al. 2011).
Sample Collection
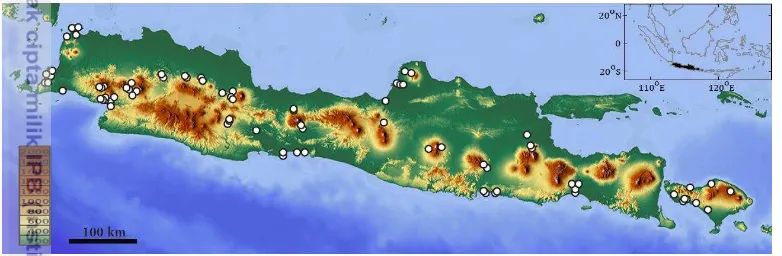
Due the high diversity of habitat and complex geological and paleocological history in the study area, this research was conducted in a total of fifty one locations across Java and Bali with the aim to explore phylogeographic patterns in South Sundaland. Three widely distributed species were selected as model: Channa gachua known as Bogo as local name (Perciformes, Channidae), Glyptothorax platypogon known as kehkel as local name (Siluriformes, Sisoridae), and Barbodes binotatus known as beunteur as local name (Cypriniformes, Cyprinidae).
4
Figure 3 Sampling locations in Java and Bali islands. One point may represent several sampling sites.
order to cover as much genetic diversity as possible and get enough statistical power to explore population genetic structure. All the specimens were photographed, individually labeled and voucher specimens were preserved in a 96% ethanol solution. Voucher specimens were deposited at the Muzeum Zoologicum Bogoriense (MZB) in the Research Centre for Biology (LIPI). DNA barcodes, photographs and collecting data were deposited on the Barcode of Life Datasystem (BOLD) in the projects Barcoding Indonesian Fishes - part VI. Phylogeography of Javanese Fish (BIFG) and Barcoding Indonesian Fishes - part VIb. Widespread primary freshwater fishes of Java and Bali (BIFGA) in the container Barcoding Indonesian Fishes of the Barcoding Fish (FishBOL) campaign. DNA barcodes were also deposited in GenBank, accessions number are available through the specimen records in BIFG and BIFGA projects
Genetic Analyses
5
Sequencing Ready Reaction” and sequencing was performed on the automatic sequencer ABI 3130 DNA Analyzer (Applied Biosystems).
Data Analyses
Phylogenetic Analyses of Gene genealogies
Haplotype phylogenies were constructed using the maximum likelihood (ML) algorithm as implemented in phyml (http://atgc.lirmm.fr/phyml) following the algorithm developed by Guindon and Gascuel (2003). The Akaike information criterion (AIC) identified the optimal model, among the 88 available, as implemented in modeltest 3.7 (Posada & Crandall 1998) and was further used for ML tree searches. Number of clades in ML haplotypes phylogenies were resolved based on Barcode Index Number (BIN) algorithm, a clustering algorithm delineating operational taxonomic unit detected through departure of genetic distances among molecular lineages (Ratnasingham & Hebert 2013). Estimation of the age of each species genealogies was inferred using the maximum pairwise divergence among haplotypes in the haplotype trees and the canonical fish substitution rate of 1% of divergence per million years (Bermingham et al. 1997).
Population Structure Analyses
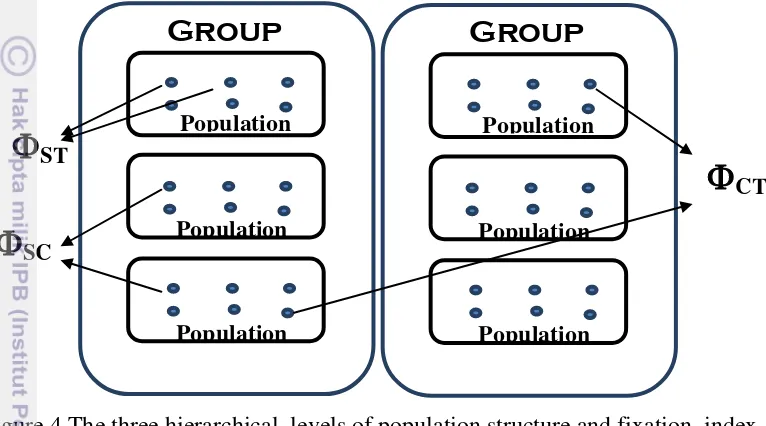
Population structure was explored through pairwise ST as implemented in Arlequin version 3.5 (Excoffier et al.2005) and significance of ST estimates was tested though permutation procedures including 1000 random replicates. Population structure was further explored through a Neighbor-Joining (NJ) tree constructed using the ape R package (Paradis et al. 2004) based on population pairwise ST estimates. A spatial analysis of molecular variance was conducted using the SAMOVA algorithm to explore the spatial partition of genetic diversity through an increasing number of genetically distinct groups of populations (Figure 3). The optimal number of groups (k) was determined in parallel based on the NJ population tree. Subsequently, the hierarchical analysis of molecular variance (AMOVA) was extracted from SAMOVA results for predefined sets of populations to infer the relative contribution of several levels of spatial partitioning of the variance including among groups (CT), among populations within groups (SC) and within populations (ST). In this hierarchical framework,
ST means genetic differentiation between individual from the same population,
SC means genetic differentiation between individual from different population in different area and CT means genetic differentiation between individual from different population and area (Figure 4). Genetic diversity within populations was estimated through several parameters including haplotype diversity, nucleotidic diversity, number of haplotypes, and polymorphic sites using the software DNAsp (Rozas et al. 2003).
6
distance (IBD) pattern in the data by plotting (1 – ) against the logarithm of geographical distance (Rousset 1997; Slatkin 1993). Isolation by Distance is a term for determining the distribution of gene frequencies over a geographic region according to the geographic distance among populations. Significance of the correlation between genetic and geographic distances was tested with the Mantel test (Legendre & Legendre 1998) as implemented in the ade4 package in R (Dray & Dufour 2007).
Figure 4 The three hierarchical levels of population structure and fixation index
RESULT
Phylogenetic Analyses
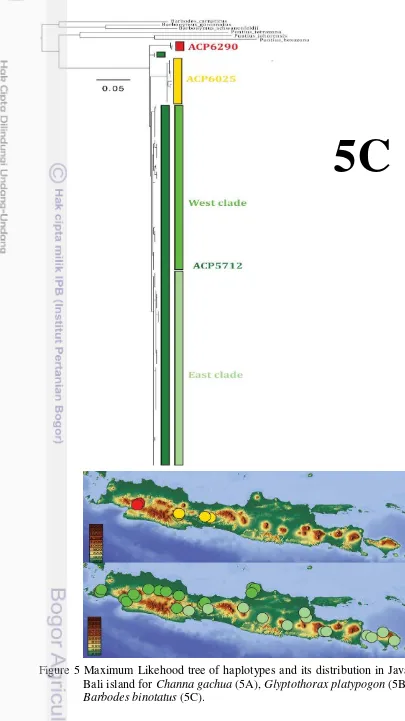
The AIC pointed to the TIM3+G model as the most likely substitution model in C. gachua. This model includes 4 base frequencies and 6 substitution categories as well as a gamma distribution accounting for differences in rates of substitution among sites. The TIM3+G most likely tree displayed a score of -lnL= 2924.20. The estimated nucleotide frequencies were A = 0.2326, C = 0.3090, G = 0.1763 and T = 0.2822, and the substitution rates were [A–C] = 2,1514, [A–G] = 24.132, [A–T] = 1.000, [C–G] = 2.1514, [C–T] = 12.6982, [G–T] = 1.0000. The BIN algorithm detected nine major clades in the tree of C. gachua (Figure 5A).
7
substitution rates [A–C] = 1.000, [A–G] = 12.1212, [A–T] = 0.1306, [C–G] = 0.1306, [C–T] = 4.6442, [G–T] = 1.0000. BIN algorithm detected seven major clades in G. platypogon tree (Figure 5B).
The AIC pointed the TPM3uf+I+G as the most likely substitution model for B. binotatus. This model includes 4 base frequencies and 6 substitution categories, the proportion of invariable sites and the gamma distribution parameter. The resulting ML score was -lnL=3233.88. The estimated nucleotide frequencies were A = 0.3023, C = 0.2763, G = 0.1404 and T=0.2809 and the substitution model incorporated the substitution rates [A–C] = 0.6322, [A–G] = 10.5781, [A–T] = 1.000, [C–G] = 0.6322, [C–T] = 10.5781, [G–T] = 1.0000. There were 3 major lineages (Figure 5C).
8
9
Figure 5 Maximum Likehood tree of haplotypes and its distribution in Java and Bali island for Channa gachua (5A), Glyptothorax platypogon (5B) and Barbodes binotatus (5C).
10
Clus ter I Cluster II
Cluster III
Cluster IV
Cluster V
Cluster I
Cluster II
Cluster III Cluster IV Cluster V
Cluster VI Cluster I
Cluster II
Cluster III
6A
6B
6C
6D
6E
6F
11
We further estimated the absolute age of the maximum divergence times for each species using the canonical 1% per million year substitution rate. In C. gachua, the maximum age estimate was 3.05 Ma (with a maximum intraspecific pairwise divergence of 6,1 %). In G. platypogon the maximum divergence was estimated to happen at 3 Ma (with a maximum intraspecific divergence of 6,0 %). In B. binotatus, the age estimate of the maximum divergence was 2.60% (with a maximum intraspecific divergence of 5.2 %).
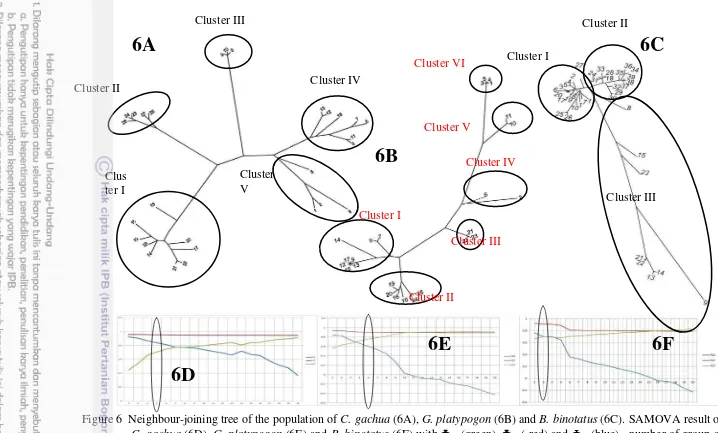
Population structure
The NJ population trees provided information about the population structure of each species. From the topology of the trees, several clusters were delineated within each species including five clusters in C. gachua (6A), six clusters in G. platypogon (6B) and three clusters in B. binotatus (6C). Below the NJ tree, the relationships between the fixation index among groups (CT), among populations within groups (SC) and within populations (ST) according to the number of population clusters indicate that among groups and among populations within group fixation index are very similar for C. gachua (6D), G. platypogon (6E) and B. binotatus (6F).
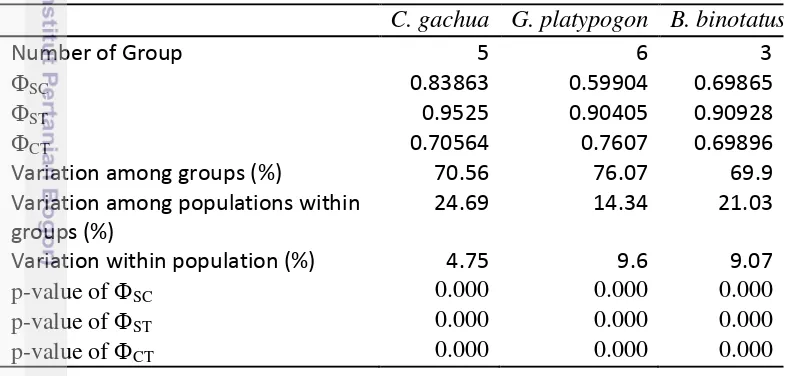
Table 1 Amova statistics and fixation indexes
Amova statistics for C. gachua (Table 1) showed high levels of structure within population (ST), among population group (SC) and among group (CT). This structure is supported by significant p-values at all levels (Table 1). Most of the variance in C. gachua, however, is explained by variations among groups (70,56%). Amova statistic of G. platypogon showed high levels of structure within population (ST), among population group (SC) and among group (CT) as evidenced by significant p-values at all levels (Table 1). Most of the variance in G. platypogon, however, is explained by variations among groups (76,07%). Amova statistics of B. binotatus showed high levels of structure within population (ST), among population group (SC) and among group (CT) as supported by significant p-values at all levels. Similarly, most of the variance in B. binotatus is explained by variations among groups (69.90%).
Variation within population (%) 4.75 9.6 9.07
p-value ofSC 0.000 0.000 0.000
p-value ofST 0.000 0.000 0.000
12
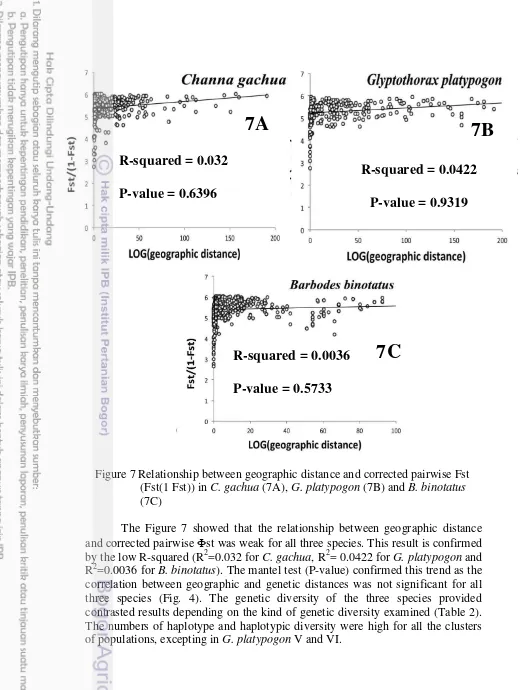
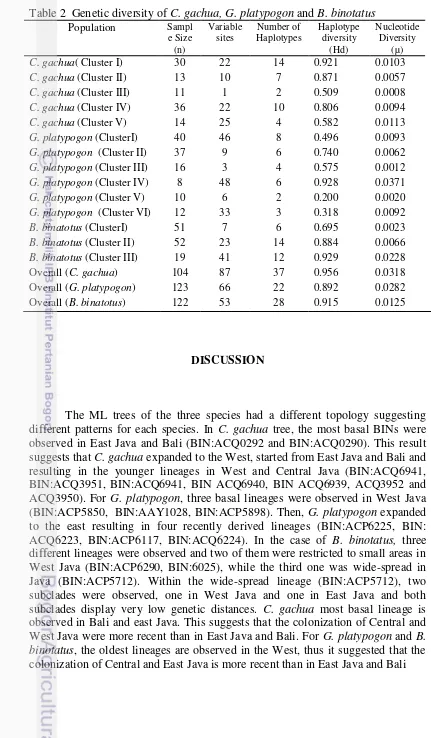
The Figure 7 showed that the relationship between geographic distance and corrected pairwise st was weak for all three species. This result is confirmed by the low R-squared (R2=0.032 for C. gachua, R2= 0.0422 for G. platypogon and R2=0.0036 for B. binotatus). The mantel test (P-value) confirmed this trend as the correlation between geographic and genetic distances was not significant for all three species (Fig. 4). The genetic diversity of the three species provided contrasted results depending on the kind of genetic diversity examined (Table 2). The numbers of haplotype and haplotypic diversity were high for all the clusters of populations, excepting in G. platypogon V and VI.
Figure 7 Relationship between geographic distance and corrected pairwise Fst (Fst(1 Fst)) in C. gachua (7A), G. platypogon (7B) and B. binotatus (7C)
7B
R-squared = 0.0422
P-value = 0.9319
7C
R-squared = 0.0036
P-value = 0.5733
7A
R-squared = 0.032
13
Table 2 Genetic diversity of C. gachua, G. platypogon and B. binotatus
14 Overall, genetic differentiation within populations, among population within group and within group was high for the three species. This result was supported by the result of the SAMOVA with the values of all tests being significant (P<0.05). It showed that the genetic structuring is high at every level for all species. In one hand, genetic variations among groups were high with overall more than 69 percent of the variation in each species located among the groups. On other hand, genetic variations within population were low according to the nucleotidic diversity. The high genetic variations among groups while low genetic variations within population were signed of an important, but recent, fragmentation of the populations as a high number of closely related haplotypes were observed. Considering that the species genealogies are estimated to originated during the late Pliocene, this observation indicates the persistence of a high number of lineages. This observation is consistent with the hypothesis that tropical biotas tend to have lower extinction rates and higher diversity because they had longer effective time for diversity to accumulate (Mittelbach et al. 2007). The high number of BIN for all species, excepting B. binotatus, is consistent with the theory. The high haplotypic diversity displayed by a larger number of population cluster also suggests that effective populations size have remained large despite the fragmentation of the populations. This was not the case, however, for two population clusters in G. platypogon (G. platypogon V and G. platypogon VI), suggesting a recent settlement of these populations.
Figure 8 Postulated distribution of land and sea in South East Asia from Hall (1998)
15
distance but a consequence of habitat fragmentation in Java and Bali landscapes. Thus, geographic distance is not sufficient to account for the distribution of genetic diversity. By contrast, a combination of vicariance events and past spatial expansions are likely to account for the high fragmentation of the populations, as observed for all the three species (Linz et al 2014). ML haplotype trees and NJ population trees provided complementary information about the evolutionary history of each of the three species.
The ML tree helped document the divergence among the haplotypes observed within population, while the NJ tree provided information about the genetic structure of the populations. Both haplotype trees and population trees are consistent with an important fragmentation of the populations. In particular, West Java populations were differentiated from East Java and Bali populations in all three species. By contrast, Central Java populations presented more contrasted population structure suggesting a dynamic of secondary contact between West and East lineages during the Pleistocene. Sea level low stand, 120 m below present level (BPL), occurred twice over the last 250.000 year BP (Voris 2000). Several rivers usually unconnected during BPL have established connections. Sea level low stands during glacial time may have enable dispersal through the temporary connection of rivers that are currently isolated physically.
CONCLUSION
16
REFERENCES
Adamson EAS. 2010. Influence of historical landscapes, drainage evolution and ecologic traits of genetic diversity in Southeast Asian freshwater snakehead fishes. PhD thesis, Queensland University of Technology. Avise JC. 2001. Phylogeography : The History and Formation of Species . USA.
Harvard
Bernatchez L, Wilson CC. 1998. Comparative phylogeography of Neartic and Paleartic fishes. Mol Ecol 7(4): 431–452.
Dray S, Dufour AB. 2007. The ade4 Package: Implementig the Duality Diagram for Ecologists. J Statis Software. 22(4) 1-20.
Dupanloup, Schneider S, Excoffier L. 2002. A simulated annealing approach to define the genetic structure of populations. Mol Ecol 11: 2571–2581. Excoffier L, Laval G, Schneider S. 2005. Arlequin ver. 3.0: An integrated
software package for population genetics data analysis. Evol Bio Info Online 1: 47–50.
Guindon S and Gascuel O. 2003.A Simple, Fast and Accurate Algorithm to Estimate Large Phylogenies by Maximum Likelihood. Syst Biol 2003 52(5): 696-704.
Hadiaty, RK. 2011. Diversitas dan hilangnya jenis-jenis ikan di Sungai Ciliwung dan Sungai Cisadane. Berita Biologi 10 (4): 491-504.
Hall R. 1998. The Plate Tectonics of Cenozoic South East Asia and the Distribution of Land and Sea. Backhuys Publishers : Leiden
Hoffman, M., C. Hilton-Taylor, A. Angulo, M. Böhm, T.M. Brooks, S. H. M. Butchart, K.E. Carpenter, J. Chanson, B. Collen & N.A. Cox. 2010. The impact of Conservation on the status of the world's vertebrates. Science 330:1503-1509.
Hebert PDN, Cywinska A, Ball SL, de Waard JR .2003. Biological identifications through DNA barcodes. Proceedings of the Royal Society of London Series B 270, 313-321.
Hubert 2008. Identifying Canadian Freshwater Fishes through DNA Barcodes. PLos One 3 : e2490
Ivanova NV, Zemlak TS, Hanner RH, Hébert PDN. 2007. Universal primers cocktails for fish DNA barcoding. Mol Ecol 7, 544-548.
Keith P, Hadiaty R, Busson F & Hubert F. 2014a. A new species of Sicyopus (Gobiidae) from Java and Bali / Une nouvelle espèce de Sicyopus (Gobiidae) d‟Indonésie.Cybium, 38 (3): 173-178.
Keith P, Hadiaty RK, Hubert N, Busson N & C Lord, 2014b. Three new species of Lentipes from Indonesia. Cybium 38(2): 133-146.
Koizumi I, Usio N, Kawai T, Azuma N, Masuda R .2012. Loss of Genetic Diversity Means Loss of Geological Information: The Endangered Japanese Crayfish Exhibits Remarkable Historical Footprints. PLoS One 7(3): e33986
17
Legendre P and Legendre L. 1998. Numerical Ecology.Amsterdam : Elsivier. Linz B, Vololonantenainab CRR, Seck A, Carod J-F, Dia D, et al. (2014)
Population Genetic Structure and Isolation by Distance of Helicobacter pylori in Senegal and Madagascar. PLoS One 9(1): e87355
Lohmann J, Bruyn M et al. 2011. Biogeography of the Indo-Australian Archipelago. Annu Rev Ecol Evol Syst 42 : 205-26.
Mittelbach G, Schemske D, Cornell H, Bush M, Harrison S, Hurlbert A, Knowlton N, Lessios H, McCain C, McCune A, McDade L, McPeek M, Near T, Price T, Ricklefs R, Roy K, Sax D, Schluter D, Sobel J and Turelli M. 2007. Evolution and The Latitudinal Diversity Gradient : Speciation, Extinction and Biogeography. Ecol Letters 10: 315–331.
Myers N, Mittermeier RA, Mittermeier CG, da Fonseca GAB, Kent J (2000) Biodiversity hotspots for conservation priorities. Nature 403, 853-858. Newman D, Pilson D. 1997. Increased probability of extinction due to decreased
genetic effective population size: experimental populations of Clarkia pulchella. J. Evol 51(2): 354–362.
Paradis E, Claude, J, Strimmer, K. 2004. APE: Analyses of Phylogenetics and492 Evolution in R language. J Bioinformatics 20: 289-290.
Posada D and Crandall KA. 1998. Modeltest : Testing the Model of DNA Substitution. J Bioinformatics.14(9):817-8.
R Development Core Team (2006) R: A Language and Environment for Statistical Computing.R Foundation for Statistical Computing, Vienna, Austria. Ratnasingham S, Hebert PDN (2013) A DNA-based registry for all animal
species: the barcode index number (BIN) system. PLoS One 8: e66213. Rousset F (1997) Genetic differentiation and estimation of gene flow from
F-Statistics under isolation by distance.genetics 145, 1219-1228.
Rozas J, Sanchez-DelBarrio JC, Messeguer X, Rozas R (2003) DnaSP, DNA polymorphism analyses by the coalescent and other methods. J Bioinformatics 19: 2496–2497.
Slatkin M (1993) Isolation by distance in equilibrium and non-equilibrium populations. J Evol 47, 264-279.
Tan MP, Jamsari AFJ, Siti Azizah MN (2012) Phylogeographic Pattern of the Striped Snakehead, Channa striata in Sundaland: Ancient River Connectivity, Geographical and Anthropogenic Singnatures. PLoS One 7(12): e52089
Voris HK. 2000. Maps of Pleistocene Sea Levels in Southeast Asia : Shorelines, River System and Time Durations. J Biogeography, 27, 1153-1167
Wang JP, Hsu KC, Chiang TY (2000) Mitochondrial DNA phylogeography of Acrossocheilus paradoxus (Cyprinidae) in Taiwan. Molecular Ecology 9(10): 1483– 1494.
APPENDIX
Table 1.Barbodes binotatus site sampling location
Site Name Province City River Lat Long BIN Process ID Catalog Num
1 BIF1510 Banten Kab Pandeglang Cisiih -6.691 105.62 BOLD:ACP5712 BIFD1071-14 MZB-BIF1510
BIF1509 Banten Kab Pandeglang Cisiih -6.691 105.62 BOLD:ACP5712 BIFD1070-14 MZB-BIF1509
BIF1511 Banten Kab Pandeglang Cisiih -6.691 105.62 BOLD:ACP5712 BIFD1072-14 MZB-BIF1511
2 BIF1531 Banten Kab Pandeglang Cijalarang -6.691 105.64 BOLD:ACP5712 BIFD1092-14 MZB-BIF1531
BIF1530 Banten Kab Pandeglang Cijalarang -6.691 105.64 BOLD:ACP5712 BIFD1091-14 MZB-BIF1530
BIF1532 Banten Kab Pandeglang Cijalarang -6.691 105.64 BOLD:ACP5712 BIFD1093-14 MZB-BIF1532
3 BIF1655 Banten Kab Pandeglang Ciseuke -6.618 105.7 BOLD:ACP5712 BIFD1216-14 MZB-BIF1655
BIF1656 Banten Kab Pandeglang Ciseuke -6.618 105.7 BOLD:ACP5712 BIFD1217-14 MZB-BIF1656
BIF1654 Banten Kab Pandeglang Ciseuke -6.618 105.7 BOLD:ACP5712 BIFD1215-14 MZB-BIF1654
4 BIF0371 West Java Kab Sukabumi Ci Pawenang -6.956 106.46 BOLD:ACP5712 BIFB346-13 MZB-BIF00371
BIF0372 West Java Kab Sukabumi Ci Pawenang -6.956 106.46 BOLD:ACP5712 BIFB347-13 MZB-BIF00372
5 BIF0202 West Java Kab Sukabumi Ci Maja -6.943 106.48 BOLD:ACP5712 BIFB177-13 MZB-BIF00202
BIF0201 West Java Kab Sukabumi Ci Maja -6.943 106.48 BOLD:ACP5712 BIFB176-13 MZB-BIF00201
6 BIF0409 West Java Kab Sukabumi Ci Haur -6.958 106.48 BOLD:ACP5712 BIFB384-13 MZB-BIF00409
BIF0408 West Java Kab Sukabumi Ci Haur -6.958 106.48 BOLD:ACP5712 BIFB383-13 MZB-BIF00408
7 BIF0110 West Java Kab Sukabumi S.Cisaat -6.773 106.77 BOLD:ACP5712 BIFB085-13 MZB-BIF00110
BIF0112 West Java Kab Sukabumi S.Cisaat -6.773 106.77 BOLD:ACP5712 BIFB087-13 MZB-BIF00112
BIF0111 West Java Kab Sukabumi S.Cisaat -6.773 106.77 BOLD:ACP5712 BIFB086-13 MZB-BIF00111
8 BIF0181 West Java Kab Sukabumi Ci Seupan -6.868 106.85 BOLD:ACP5712 BIFB156-13 MZB-BIF00181
9 BIF0166 West Java Kab Sukabumi Ci Heulang -6.838 106.89 BOLD:ACQ3951 BIFB131-13 MZB-BIF00166
BIF0165 West Java Kab Sukabumi Ci Heulang -6.838 106.89 BOLD:ACQ3951 BIFB130-13 MZB-BIF00165
10 BIF0441 West Java Kab Purwakarta Ci Nantke -6.528 107.41 BOLD:ACP5712 BIFD002-13 MZB-BIF00441
BIF0440 West Java Kab Purwakarta Ci Nantke -6.528 107.41 BOLD:ACP5712 BIFD001-13 MZB-BIF00440
BIF0446 West Java Kab Purwakarta Ci Nantke -6.528 107.41 BOLD:ACP5712 BIFD007-13 MZB-BIF00446
11 BIF0491 West Java Kab Purwakarta Ci Asem -6.553 107.67 BOLD:ACP5712 BIFD052-13 MZB-BIF00491
12 BIF0556 West Java Kab Purwakarta Ci Juray -6.609 107.85 BOLD:ACP5712 BIFD117-13 MZB-BIF00556
BIF0554 West Java Kab Purwakarta Ci Juray -6.609 107.85 BOLD:ACP5712 BIFD115-13 MZB-BIF00554
BIF0555 West Java Kab Purwakarta Ci Juray -6.609 107.85 BOLD:ACP5712 BIFD116-13 MZB-BIF00555
13 BIF0642 West Java Kab Tasikmalaya Ci Gede -7.284 108.2 BOLD:ACP6025 BIFD203-13 MZB-BIF00642
BIF0641 West Java Kab Tasikmalaya Ci Gede -7.284 108.2 BOLD:ACP6025 BIFD202-13 MZB-BIF00641
14 BIF0617 West Java Kab Tasikmalaya Ci Baruay -7.251 108.22 BOLD:ACP6025 BIFD178-13 MZB-BIF00617
BIF0616 West Java Kab Tasikmalaya Ci Baruay -7.251 108.22 BOLD:ACP6025 BIFD177-13 MZB-BIF00616
15 BIF0602 West Java Kab Tasikmalaya Ci Galugur -7.261 108.22 BOLD:ACP5712 BIFD163-13 MZB-BIF00602
BIF0601 West Java Kab Tasikmalaya Ci Galugur -7.261 108.22 BOLD:ACP5712 BIFD162-13 MZB-BIF00601
BIF0600 West Java Kab Tasikmalaya Ci Galugur -7.261 108.22 BOLD:ACP5712 BIFD161-13 MZB-BIF00600
16 BIF0705 West Java Kab Majalenka Ci Longkrang -6.895 108.3 BOLD:ACP5712 BIFD266-13 MZB-BIF00705
BIF0704 West Java Kab Majalenka Ci Longkrang -6.895 108.3 BOLD:ACP5712 BIFD265-13 MZB-BIF00704
BIF0703 West Java Kab Majalenka Ci Longkrang -6.892 108.3 BOLD:ACP5712 BIFD264-13 MZB-BIF00703
17 BIF0737 West Java Kab Majalenka Ci Beber -6.845 108.31 BOLD:ACP5712 BIFD298-13 MZB-BIF00737
BIF0738 West Java Kab Majalenka Ci Beber -6.847 108.31 BOLD:ACP5712 BIFD299-13 MZB-BIF00738
BIF0739 West Java Kab Majalenka Ci Beber -6.847 108.31 BOLD:ACP5712 BIFD300-13 MZB-BIF00739
18 BIF0672 West Java Kab Camis Ci Hapitan -7.41 108.44 BOLD:ACP5712 BIFD233-13 MZB-BIF00672
BIF0674 West Java Kab Camis Ci Hapitan -7.41 108.44 BOLD:ACP5712 BIFD235-13 MZB-BIF00674
BIF0673 West Java Kab Camis Ci Hapitan -7.41 108.44 BOLD:ACP5712 BIFD234-13 MZB-BIF00673
BIF0933 Central Java Kab Tegal Kali Timbang -7.16 109.15 BOLD:ACP5712 BIFD494-13 MZB-BIF00933
BIF0934 Central Java Kab Tegal Kali Timbang -7.16 109.15 BOLD:ACP5712 BIFD495-13 MZB-BIF00934
20 BIF0908 Central Java Kab Tegal Kali Longkrang -7.16 109.16 BOLD:ACP5712 BIFD469-13 MZB-BIF00908
BIF0909 Central Java Kab Tegal Kali Longkrang -7.16 109.16 BOLD:ACP5712 BIFD470-13 MZB-BIF00909
21 BIF1131 Central Java Kab Temanggung Fish market -7.336 109.18 BOLD:ACP6025 BIFD692-13 MZB-BIF01131
22 BIF1123 Central Java Kab Temanggung Kali Cabe -7.33 109.19 BOLD:ACP6025 BIFD684-13 MZB-BIF01123
BIF1124 Central Java Kab Temanggung Kali Cabe -7.33 109.19 BOLD:ACP6025 BIFD685-13 MZB-BIF01124
BIF1125 Central Java Kab Temanggung Kali Cabe -7.33 109.19 BOLD:ACP6025 BIFD686-13 MZB-BIF01125
23 BIF0881 Central Java Kab Purwokerto Kali Pelus -7.371 109.23 BOLD:ACP6025 BIFD442-13 MZB-BIF00881
BIF0860 Central Java Kab Purwokerto Kali Pelus -7.371 109.23 BOLD:ACP5712 BIFD421-13 MZB-BIF00860
BIF0884 Central Java Kab Purwokerto Kali Pelus -7.371 109.23 BOLD:ACP6025 BIFD445-13 MZB-BIF00884
BIF0859 Central Java Kab Purwokerto Kali Pelus -7.371 109.23 BOLD:ACP5712 BIFD420-13 MZB-BIF00859
24 BIF1229 Central Java Kab Ambarawa Danau Rawa
Pening
-7.27 110.42 BOLD:ACP5712 BIFD790-13 MZB-BIF01229
BIF1231 Central Java Kab Ambarawa Danau Rawa
Pening
-7.27 110.42 BOLD:ACP5712 BIFD792-13 MZB-BIF01231
BIF1230 Central Java Kab Ambarawa Danau Rawa
Pening
-7.27 110.42 BOLD:ACP5712 BIFD791-13 MZB-BIF01230
BIF1232 Central Java Kab Ambarawa Danau Rawa
Pening
-7.27 110.42 BOLD:ACP5712 BIFD793-13 MZB-BIF01232
BIF1233 Central Java Kab Ambarawa Danau Rawa
Pening
-7.27 110.42 BOLD:ACP5712 BIFD794-13 MZB-BIF01233
25 BIF3481 Central Java Kab Japara Kali Gede -6.608 110.73 BOLD:ACP5712 BIFD2602-15 MZB-BIF3481
BIF3480 Central Java Kab Japara Kali Gede -6.608 110.73 BOLD:ACP5712 BIFD2601-15 MZB-BIF3480
BIF3483 Central Java Kab Japara Kali Gede -6.608 110.73 BOLD:ACP5712 BIFD2604-15 MZB-BIF3483
BIF3482 Central Java Kab Japara Kali Gede -6.608 110.73 BOLD:ACP5712 BIFD2603-15 MZB-BIF3482
26 BIF3445 Central Java Kab Japara Bakalan -6.73 110.72 BOLD:ACP5712 BIFD2566-15 MZB-BIF3445
BIF3446 Central Java Kab Japara Bakalan -6.73 110.72 BOLD:ACP5712 BIFD2567-15 MZB-BIF3446
BIF3447 Central Java Kab Japara Bakalan -6.73 110.72 BOLD:ACP5712 BIFD2568-15 MZB-BIF3447
27 BIF3511 Central Java Kab Japara Kali Salak -6.62 110.81 BOLD:ACP5712 BIFD2632-15 MZB-BIF3511
BIF3508 Central Java Kab Japara Kali Salak -6.62 110.81 BOLD:ACP5712 BIFD2629-15 MZB-BIF3508
BIF3509 Central Java Kab Japara Kali Salak -6.62 110.81 BOLD:ACP5712 BIFD2630-15 MZB-BIF3509
BIF3510 Central Java Kab Japara Kali Salak -6.62 110.81 BOLD:ACP5712 BIFD2631-15 MZB-BIF3510
28 BIF1814 Central Java Kab Karang Anyar Samin -7.654 111.08 BOLD:ACP5712 BIFD1375-14 MZB-BIF1814
BIF1815 Central Java Kab Karang Anyar Samin -7.654 111.08 BOLD:ACP5712 BIFD1376-14 MZB-BIF1815
BIF1812 Central Java Kab Karang Anyar Samin -7.654 111.08 BOLD:ACP5712 BIFD1373-14 MZB-BIF1812
BIF1813 Central Java Kab Karang Anyar Samin -7.654 111.08 BOLD:ACP5712 BIFD1374-14 MZB-BIF1813
29 BIF1888 Central Java Kab Kediri Bruni -7.915 111.95 BOLD:ACP5712 BIFD1449-14 MZB-BIF1888
BIF1890 Central Java Kab Kediri Bruni -7.915 111.95 BOLD:ACP5712 BIFD1451-14 MZB-BIF1890
BIF1889 Central Java Kab Kediri Bruni -7.915 111.95 BOLD:ACP5712 BIFD1450-14 MZB-BIF1889
BIF1891 Central Java Kab Kediri Bruni -7.915 111.95 BOLD:ACP5712 BIFD1452-14 MZB-BIF1891
30 BIF1993 East Java Kab Blitar Ngerjo -8.269 112.05 BOLD:ACP5712 BIFD1554-14 MZB-BIF1993
BIF1995 East Java Kab Blitar Ngerjo -8.269 112.05 BOLD:ACP5712 BIFD1556-14 MZB-BIF1995
BIF1992 East Java Kab Blitar Ngerjo -8.269 112.05 BOLD:ACP5712 BIFD1553-14 MZB-BIF1992
BIF1994 East Java Kab Blitar Ngerjo -8.269 112.05 BOLD:ACP5712 BIFD1555-14 MZB-BIF1994
31 BIF3681 East Java Kab Mojokerto Pasar Ikan
Mojokerto
-7.443 112.46 BOLD:ACP5712 BIFD2801-15 MZB-BIF3681
BIF3680 East Java Kab Mojokerto Pasar Ikan
Mojokerto
-7.443 112.4 BOLD:ACP5712 BIFD2800-15 MZB-BIF3680
BIF3679 East Java Kab Mojokerto Pasar Ikan
Mojokerto
-7.443 112.4 BOLD:ACP5712 BIFD2799-15 MZB-BIF3679
BIF3678 East Java Kab Mojokerto Pasar Ikan
Mojokerto
-7.443 112.4 BOLD:ACP5712 BIFD2798-15 MZB-BIF3678
32 BIF3605 East Java Kab Mojokerto Kali Dauwan -7.665 112.51 BOLD:ACP5712 BIFD2726-15 MZB-BIF3605
BIF3602 East Java Kab Mojokerto Kali Dauwan -7.665 112.51 BOLD:ACP5712 BIFD2723-15 MZB-BIF3602
BIF3604 East Java Kab Mojokerto Kali Dauwan -7.665 112.51 BOLD:ACP5712 BIFD2725-15 MZB-BIF3604
33 BIF3577 East Java Kab Mojokerto Kali Kromong -7.669 112.53 BOLD:ACP5712 BIFD2698-15 MZB-BIF3577
BIF3580 East Java Kab Mojokerto Kali Kromong -7.669 112.53 BOLD:ACP5712 BIFD2701-15 MZB-BIF3580
BIF3581 East Java Kab Mojokerto Kali Kromong -7.669 112.53 BOLD:ACP5712 BIFD2702-15 MZB-BIF3581
BIF3578 East Java Kab Mojokerto Kali Kromong -7.669 112.53 BOLD:ACP5712 BIFD2699-15 MZB-BIF3578
BIF3579 East Java Kab Mojokerto Kali Kromong -7.669 112.53 BOLD:ACP5712 BIFD2700-15 MZB-BIF3579
34 BIF2183 East Java Kab Lumajang Mujur -8.273 113.17 BOLD:ACP5712 BIFD1744-14 MZB-BIF2183
BIF2184 East Java Kab Lumajang Mujur -8.273 113.17 BOLD:ACP5712 BIFD1745-14 MZB-BIF2184
BIF2186 East Java Kab Lumajang Mujur -8.273 113.17 BOLD:ACP5712 BIFD1747-14 MZB-BIF2186
BIF2185 East Java Kab Lumajang Mujur -8.273 113.17 BOLD:ACP5712 BIFD1746-14 MZB-BIF2185
BIF2182 East Java Kab Lumajang Mujur -8.273 113.17 BOLD:ACP5712 BIFD1743-14 MZB-BIF2182
35 BIF2115 East Java Kab Lumajang Asem -8.131 113.22 BOLD:ACP5712 BIFD1676-14 MZB-BIF2115
BIF2114 East Java Kab Lumajang Asem -8.131 113.22 BOLD:ACP5712 BIFD1675-14 MZB-BIF2114
BIF2117 East Java Kab Lumajang Asem -8.131 113.22 BOLD:ACP5712 BIFD1678-14 MZB-BIF2117
BIF2116 East Java Kab Lumajang Asem -8.131 113.22 BOLD:ACP5712 BIFD1677-14 MZB-BIF2116
BIF2113 East Java Kab Lumajang Asem -8.131 113.22 BOLD:ACP5712 BIFD1674-14 MZB-BIF2113
36 BIF2480 West Bali Kab Jembrana Bendung Bekel -8.283 114.6 BOLD:ACP5712 BIFD2040-14 MZB-BIF2480
BIF2482 West Bali Kab Jembrana Bendung Bekel -8.283 114.6 BOLD:ACP5712 BIFD2042-14 MZB-BIF2482
BIF2481 West Bali Kab Jembrana Bendung Bekel -8.283 114.6 BOLD:ACP5712 BIFD2041-14 MZB-BIF2481
37 BIF2464 West Bali Kab Jembrana Yeh Sumbul -8.36 114.78 BOLD:ACP5712 BIFD2024-14 MZB-BIF2464
BIF2463 West Bali Kab Jembrana Yeh Sumbul -8.36 114.78 BOLD:ACP5712 BIFD2023-14 MZB-BIF2463
38 BIF2725 West Bali Kab Kelungkung Tukad Unda BOLD:ACP5712 BIFD2285-14 MZB-BIF2725
BIF2723 West Bali Kab Kelungkung Tukad Unda BOLD:ACP5712 BIFD2283-14 MZB-BIF2723
BIF2724 West Bali Kab Kelungkung Tukad Unda BOLD:ACP5712 BIFD2284-14 MZB-BIF2724
BIF2915 West Bali Kab Buleleng Danau Buyan BOLD:ACP5712 BIFD2475-14 MZB-BIF2915
BIF2925 West Bali Kab Buleleng Danau Buyan BOLD:ACP5712 BIFD2485-14 MZB-BIF2925
BIF2916 West Bali Kab Buleleng Danau Buyan BOLD:ACP5712 BIFD2476-14 MZB-BIF2916
Table 2.Channa gachua site sampling location
No. Name Province City River Lat Long BIN Process ID Catalog Num
1 BIF1650 Banten Kab Lebak Cibareno -6.96 105.40 BOLD:AAY1028 BIFD1211-14 MZB-BIF1650
2 BIF1787 West Java Kab Sukabumi Ciawi -6.84 106.43 BOLD:AAY1028 BIFD1348-14 MZB-BIF1787
BIF1788 West Java Kab Sukabumi Ciawi -6.84 106.43 BOLD:AAY1028 BIFD1349-14 MZB-BIF1788
BIF1786 West Java Kab Sukabumi Ciawi -6.84 106.43 BOLD:AAY1028 BIFD1347-14 MZB-BIF1786
3 BIF0107 West Java Kab Sukabumi S.Cisaat -6.77 106.77 BOLD:AAY1028 BIFB082-13 MZB-BIF00107
BIF0108 West Java Kab Sukabumi S.Cisaat -6.77 106.77 BOLD:AAY1028 BIFB083-13 MZB-BIF00108
4 BIF0176 West Java Kab Sukabumi Ci Seupan -6.87 106.86 BOLD:AAY1028 BIFB151-13 MZB-BIF00176
BIF0177 West Java Kab Sukabumi Ci Seupan -6.87 106.86 BOLD:AAY1028 BIFB152-13 MZB-BIF00177
BIF0179 West Java Kab Sukabumi Ci Seupan -6.87 106.86 BOLD:AAY1028 BIFB154-13 MZB-BIF00179
BIF0178 West Java Kab Sukabumi Ci Seupan -6.87 106.86 BOLD:AAY1028 BIFB153-13 MZB-BIF00178
5 BIF0156 West Java Kab Sukabumi Ci Heulang -6.84 106.90 BOLD:AAY1028 BIFB131-13 MZB-BIF00156
BIF0157 West Java Kab Sukabumi Ci Heulang -6.84 106.90 BOLD:AAY1028 BIFB132-13 MZB-BIF00157
BIF0159 West Java Kab Sukabumi Ci Heulang -6.84 106.90 BOLD:AAY1028 BIFB134-13 MZB-BIF00159
BIF0158 West Java Kab Sukabumi Ci Heulang -6.84 106.90 BOLD:AAY1028 BIFB133-13 MZB-BIF00158
6 BIF0498 West Java Kab Purwakarta Ci Asem -6.56 107.68 BOLD:ACP6117 BIFD059-13 MZB-BIF00498
BIF0496 West Java Kab Purwakarta Ci Asem -6.56 107.68 BOLD:ACP5850 BIFD057-13 MZB-BIF00496
BIF0497 West Java Kab Purwakarta Ci Asem -6.56 107.68 BOLD:ACP5850 BIFD058-13 MZB-BIF00497
7 BIF0567 West Java Kab Purwakarta Ci Juray -6.61 107.86 BOLD:ACP6117 BIFD128-13 MZB-BIF00567
BIF0564 West Java Kab Purwakarta Ci Juray -6.61 107.86 BOLD:ACP6117 BIFD125-13 MZB-BIF00564
BIF0568 West Java Kab Purwakarta Ci Juray -6.61 107.86 BOLD:ACP6117 BIFD129-13 MZB-BIF00568
BIF0565 West Java Kab Purwakarta Ci Juray -6.61 107.86 BOLD:ACP6117 BIFD126-13 MZB-BIF00565
BIF0566 West Java Kab Purwakarta Ci Juray -6.61 107.86 BOLD:ACP6117 BIFD127-13 MZB-BIF00566
8 BIF0525 West Java Kab Purwakarta Ci Panagara -6.60 107.86 BOLD:ACP6117 BIFD086-13 MZB-BIF00525
9 BIF0779 West Java Kab Majalenka Ci Lutung -6.83 108.16 BOLD:ACP6224 BIFD340-13 MZB-BIF00779
BIF0782 West Java Kab Majalenka Ci Lutung -6.83 108.16 BOLD:ACP6224 BIFD343-13 MZB-BIF00782
BIF0780 West Java Kab Majalenka Ci Lutung -6.83 108.16 BOLD:ACP6224 BIFD341-13 MZB-BIF00780
BIF0781 West Java Kab Majalenka Ci Lutung -6.83 108.16 BOLD:ACP6224 BIFD342-13 MZB-BIF00781
BIF0783 West Java Kab Majalenka Ci Lutung -6.83 108.16 BOLD:ACP6224 BIFD344-13 MZB-BIF00783
10 BIF0664 West Java Kab Tasikmalaya Ci Gede -7.28 108.21 BOLD:ACP5898 BIFD225-13 MZB-BIF00664
BIF0661 West Java Kab Tasikmalaya Ci Gede -7.28 108.21 BOLD:ACP5898 BIFD222-13 MZB-BIF00661
BIF0665 West Java Kab Tasikmalaya Ci Gede -7.28 108.21 BOLD:ACP5898 BIFD226-13 MZB-BIF00665
BIF0663 West Java Kab Tasikmalaya Ci Gede -7.28 108.21 BOLD:ACP5898 BIFD224-13 MZB-BIF00663
BIF0662 West Java Kab Tasikmalaya Ci Gede -7.28 108.21 BOLD:ACP5898 BIFD223-13 MZB-BIF00662
11 BIF0583 West Java Kab Tasikmalaya Ci Galugur -7.26 108.22 BOLD:ACP5898 BIFD144-13 MZB-BIF00583
BIF0585 West Java Kab Tasikmalaya Ci Galugur -7.26 108.22 BOLD:ACP5898 BIFD146-13 MZB-BIF 585
BIF0584 West Java Kab Tasikmalaya Ci Galugur -7.26 108.22 BOLD:ACP5898 BIFD145-13 MZB-BIF 584
BIF0586 West Java Kab Tasikmalaya Ci Galugur -7.26 108.22 BOLD:ACP5898 BIFD147-13 MZB-BIF 586
BIF0582 West Java Kab Tasikmalaya Ci Galugur -7.26 108.22 BOLD:ACP5898 BIFD143-13 MZB-BIF 582
12 BIF0715 West Java Kab Majalenka Ci Longkrang -6.90 108.31 BOLD:ACP6224 BIFD276-13 MZB-BIF 715
BIF0717 West Java Kab Majalenka Ci Longkrang -6.90 108.31 BOLD:ACP6224 BIFD278-13 MZB-BIF 717
BIF0713 West Java Kab Majalenka Ci Longkrang -6.90 108.31 BOLD:ACP6224 BIFD274-13 MZB-BIF00713
BIF0716 West Java Kab Majalenka Ci Longkrang -6.90 108.31 BOLD:ACP6224 BIFD277-13 MZB-BIF00716
13 BIF0743 West Java Kab Majalenka Ci Beber -6.85 108.31 BOLD:ACP6224 BIFD304-13 MZB-BIF00743
BIF0742 West Java Kab Majalenka Ci Beber -6.85 108.31 BOLD:ACP6224 BIFD303-13 MZB-BIF00742
BIF0746 West Java Kab Majalenka Ci Beber -6.85 108.31 BOLD:ACP6224 BIFD307-13 MZB-BIF00746
BIF0745 West Java Kab Majalenka Ci Beber -6.85 108.31 BOLD:ACP6224 BIFD306-13 MZB-BIF00745
BIF0744 West Java Kab Majalenka Ci Beber -6.85 108.31 BOLD:ACP6224 BIFD305-13 MZB-BIF00744
14 BIF0928 Central Java Kab Tegal Kali Timbang -7.17 109.16 BOLD:ACP6224 BIFD489-13 MZB-BIF00928
BIF0930 Central Java Kab Tegal Kali Timbang -7.17 109.16 BOLD:ACP6224 BIFD491-13 MZB-BIF00930
BIF0931 Central Java Kab Tegal Kali Timbang -7.17 109.16 BOLD:ACP6224 BIFD492-13 MZB-BIF00931
BIF0927 Central Java Kab Tegal Kali Timbang -7.17 109.16 BOLD:ACP6224 BIFD488-13 MZB-BIF00927
BIF0932 Central Java Kab Tegal Kali Timbang -7.17 109.16 BOLD:ACP6224 BIFD493-13 MZB-BIF00932
BIF0929 Central Java Kab Tegal Kali Timbang -7.17 109.16 BOLD:ACP6224 BIFD490-13 MZB-BIF00929
15 BIF0900 Central Java Kab Tegal Kali Longkrang -7.17 109.16 BOLD:ACP6224 BIFD461-13 MZB-BIF00900
BIF0897 Central Java Kab Tegal Kali Longkrang -7.17 109.16 BOLD:ACP6224 BIFD458-13 MZB-BIF00897
BIF0901 Central Java Kab Tegal Kali Longkrang -7.17 109.16 BOLD:ACP6224 BIFD462-13 MZB-BIF00901
BIF0902 Central Java Kab Tegal Kali Longkrang -7.17 109.16 BOLD:ACP6224 BIFD463-13 MZB-BIF00902
BIF0904 Central Java Kab Tegal Kali Longkrang -7.17 109.16 BOLD:ACP6224 BIFD465-13 MZB-BIF00904
BIF0899 Central Java Kab Tegal Kali Longkrang -7.17 109.16 BOLD:ACP6224 BIFD460-13 MZB-BIF00899
BIF0903 Central Java Kab Tegal Kali Longkrang -7.17 109.16 BOLD:ACP6224 BIFD464-13 MZB-BIF00903
BIF0898 Central Java Kab Tegal Kali Longkrang -7.17 109.16 BOLD:ACP6224 BIFD459-13 MZB-BIF00898
16 BIF1099 Central Java Kab Temanggung Kali Cabe -7.34 109.19 BOLD:ACP6223 BIFD660-13 MZB-BIF01099
BIF1097 Central Java Kab Temanggung Kali Cabe -7.34 109.19 BOLD:ACP6223 BIFD658-13 MZB-BIF01097
BIF1101 Central Java Kab Temanggung Kali Cabe -7.34 109.19 BOLD:ACP6223 BIFD662-13 MZB-BIF01101
BIF1104 Central Java Kab Temanggung Kali Cabe -7.34 109.19 BOLD:ACP6223 BIFD665-13 MZB-BIF01104
BIF1103 Central Java Kab Temanggung Kali Cabe -7.34 109.19 BOLD:ACP6223 BIFD664-13 MZB-BIF01103
BIF1102 Central Java Kab Temanggung Kali Cabe -7.34 109.19 BOLD:ACP6223 BIFD663-13 MZB-BIF01102
17 BIF0846 Central Java Kab Purwokerto Kali Pelus -7.37 109.24 BOLD:ACP6224 BIFD407-13 MZB-BIF00846
BIF0843 Central Java Kab Purwokerto Kali Pelus -7.37 109.24 BOLD:ACP6224 BIFD404-13 MZB-BIF00843
BIF0841 Central Java Kab Purwokerto Kali Pelus -7.37 109.24 BOLD:ACP6224 BIFD402-13 MZB-BIF00841
BIF0847 Central Java Kab Purwokerto Kali Pelus -7.37 109.24 BOLD:ACP6224 BIFD408-13 MZB-BIF00847
BIF0845 Central Java Kab Purwokerto Kali Pelus -7.37 109.24 BOLD:ACP6224 BIFD406-13 MZB-BIF00845
18 BIF3522 Central Java Kab Japara Kali Salak -6.62 110.81 BOLD:ACP6223 BIFD2643-15 MZB-BIF3522
BIF3523 Central Java Kab Japara Kali Salak -6.62 110.81 BOLD:ACP6223 BIFD2644-15 MZB-BIF3523
BIF3524 Central Java Kab Japara Kali Salak -6.62 110.81 BOLD:ACP6223 BIFD2645-15 MZB-BIF3524
BIF3525 Central Java Kab Japara Kali Salak -6.62 110.81 BOLD:ACP6223 BIFD2646-15 MZB-BIF3525
19 BIF1849 Central Java Kab Karang Anyar Samin -7.65 111.08 BOLD:ACP6223 BIFD1410-14 MZB-BIF1849
BIF1844 Central Java Kab Karang Anyar Samin -7.65 111.08 BOLD:ACP6223 BIFD1405-14 MZB-BIF1844
BIF1846 Central Java Kab Karang Anyar Samin -7.65 111.08 BOLD:ACP6223 BIFD1407-14 MZB-BIF1846
BIF1847 Central Java Kab Karang Anyar Samin -7.65 111.08 BOLD:ACP6223 BIFD1408-14 MZB-BIF1847
BIF1851 Central Java Kab Karang Anyar Samin -7.65 111.08 BOLD:ACP6223 BIFD1412-14 MZB-BIF1851
BIF1848 Central Java Kab Karang Anyar Samin -7.65 111.08 BOLD:ACP6223 BIFD1409-14 MZB-BIF1848
BIF1850 Central Java Kab Karang Anyar Samin -7.65 111.08 BOLD:ACP6223 BIFD1411-14 MZB-BIF1850
BIF1845 Central Java Kab Karang Anyar Samin -7.65 111.08 BOLD:ACP6223 BIFD1406-14 MZB-BIF1845
20 BIF1868 Central Java Kab Magetan Gema -7.62 111.30 BOLD:ACP6223 BIFD1429-14 MZB-BIF1868
BIF1866 Central Java Kab Magetan Gema -7.62 111.30 BOLD:ACP6223 BIFD1427-14 MZB-BIF1866
BIF1867 Central Java Kab Magetan Gema -7.62 111.30 BOLD:ACP6223 BIFD1428-14 MZB-BIF1867
BIF1865 Central Java Kab Magetan Gema -7.62 111.30 BOLD:ACP6223 BIFD1426-14 MZB-BIF1865
BIF1863 Central Java Kab Magetan Gema -7.62 111.30 BOLD:ACP6223 BIFD1424-14 MZB-BIF1863
21 BIF1926 East Java Kab Kediri Bruni -7.89 111.90 BOLD:ACP6225 BIFD1487-14 MZB-BIF1926
BIF1925 East Java Kab Kediri Bruni -7.89 111.90 BOLD:ACP6225 BIFD1486-14 MZB-BIF1925
BIF1928 East Java Kab Kediri Bruni -7.89 111.90 BOLD:ACP6225 BIFD1489-14 MZB-BIF1928
BIF1924 East Java Kab Kediri Bruni -7.89 111.90 BOLD:ACP6225 BIFD1485-14 MZB-BIF1924
BIF1927 East Java Kab Kediri Bruni -7.89 111.90 BOLD:ACP6225 BIFD1488-14 MZB-BIF1927
BIF1923 East Java Kab Kediri Bruni -7.89 111.90 BOLD:ACP6225 BIFD1484-14 MZB-BIF1923
BIF1921 East Java Kab Kediri Bruni -7.89 111.90 BOLD:ACP6225 BIFD1482-14 MZB-BIF1921
BIF1922 East Java Kab Kediri Bruni -7.89 111.90 BOLD:ACP6225 BIFD1483-14 MZB-BIF1922
22 BIF3631 East Java Kab Mojokerto Kali Dauwan -7.67 112.51 BOLD:ACP6225 BIFD2751-15 MZB-BIF3631
BIF3633 East Java Kab Mojokerto Kali Dauwan -7.67 112.51 BOLD:ACP6225 BIFD2753-15 MZB-BIF3633
BIF3632 East Java Kab Mojokerto Kali Dauwan -7.67 112.51 BOLD:ACP6225 BIFD2752-15 MZB-BIF3632
BIF3630 East Java Kab Mojokerto Kali Dauwan -7.67 112.51 BOLD:ACP6225 BIFD2750-15 MZB-BIF3630
23 BIF3591 East Java Kab Mojokerto Kali Kromong -7.67 112.53 BOLD:ACP6225 BIFD2712-15 MZB-BIF3591
BIF3593 East Java Kab Mojokerto Kali Kromong -7.67 112.53 BOLD:ACP6225 BIFD2714-15 MZB-BIF3593
BIF3592 East Java Kab Mojokerto Kali Kromong -7.67 112.53 BOLD:ACP6225 BIFD2713-15 MZB-BIF3592
BIF3590 East Java Kab Mojokerto Kali Kromong -7.67 112.53 BOLD:ACP6225 BIFD2711-15 MZB-BIF3590
24 BIF2090 East Java Kab Lumajang Bicoro -8.13 113.11 BOLD:ACP6223 BIFD1651-14 MZB-BIF2090
BIF2092 East Java Kab Lumajang Bicoro -8.13 113.11 BOLD:ACP6223 BIFD1653-14 MZB-BIF2092
BIF2093 East Java Kab Lumajang Bicoro -8.13 113.11 BOLD:ACP6223 BIFD1654-14 MZB-BIF2093
BIF2095 East Java Kab Lumajang Bicoro -8.13 113.11 BOLD:ACP6223 BIFD1656-14 MZB-BIF2095
BIF2091 East Java Kab Lumajang Bicoro -8.13 113.11 BOLD:ACP6223 BIFD1652-14 MZB-BIF2091
BIF2088 East Java Kab Lumajang Bicoro -8.13 113.11 BOLD:ACP6223 BIFD1649-14 MZB-BIF2088
BIF2089 East Java Kab Lumajang Bicoro -8.13 113.11 BOLD:ACP6223 BIFD1650-14 MZB-BIF2089
BIF2094 East Java Kab Lumajang Bicoro -8.13 113.11 BOLD:ACP6223 BIFD1655-14 MZB-BIF2094
BIF2128 East Java Kab Lumajang Asem -8.13 113.23 BOLD:ACP6223 BIFD1689-14 MZB-BIF2128
BIF2133 East Java Kab Lumajang Asem -8.13 113.23 BOLD:ACP6223 BIFD1694-14 MZB-BIF2133
BIF2134 East Java Kab Lumajang Asem -8.13 113.23 BOLD:ACP6223 BIFD1695-14 MZB-BIF2134
BIF2129 East Java Kab Lumajang Asem -8.13 113.23 BOLD:ACP6223 BIFD1690-14 MZB-BIF2129
BIF2132 East Java Kab Lumajang Asem -8.13 113.23 BOLD:ACP6223 BIFD1693-14 MZB-BIF2132
Table 3.Glyptothorax platypogon site sampling location
Site Name Province City River Lat Long BIN Process ID Catalog Num
1 BIF1650 Banten Kab Lebak Cibareno -6.96333 105.395 BOLD:AAY1028 BIFD1211-14 MZB-BIF1650
2 BIF1787 West Java Kab Sukabumi Ciawi -6.83944 106.434 BOLD:AAY1028 BIFD1348-14 MZB-BIF1787
BIF1788 West Java Kab Sukabumi Ciawi -6.83944 106.434 BOLD:AAY1028 BIFD1349-14 MZB-BIF1788
BIF1786 West Java Kab Sukabumi Ciawi -6.83944 106.434 BOLD:AAY1028 BIFD1347-14 MZB-BIF1786
3 BIF0107 West Java Kab Sukabumi S.Cisaat -6.773 106.773 BOLD:AAY1028 BIFB082-13 MZB-BIF00107
BIF0108 West Java Kab Sukabumi S.Cisaat -6.773 106.773 BOLD:AAY1028 BIFB083-13 MZB-BIF00108
4 BIF0176 West Java Kab Sukabumi Ci Seupan -6.868 106.858 BOLD:AAY1028 BIFB151-13 MZB-BIF00176
BIF0177 West Java Kab Sukabumi Ci Seupan -6.868 106.858 BOLD:AAY1028 BIFB152-13 MZB-BIF00177
BIF0179 West Java Kab Sukabumi Ci Seupan -6.868 106.858 BOLD:AAY1028 BIFB154-13 MZB-BIF00179
BIF0178 West Java Kab Sukabumi Ci Seupan -6.868 106.858 BOLD:AAY1028 BIFB153-13 MZB-BIF00178
5 BIF0156 West Java Kab Sukabumi Ci Heulang -6.838 106.896 BOLD:AAY1028 BIFB131-13 MZB-BIF00156
BIF0157 West Java Kab Sukabumi Ci Heulang -6.838 106.896 BOLD:AAY1028 BIFB132-13 MZB-BIF00157
BIF0159 West Java Kab Sukabumi Ci Heulang -6.838 106.896 BOLD:AAY1028 BIFB134-13 MZB-BIF00159
BIF0158 West Java Kab Sukabumi Ci Heulang -6.838 106.896 BOLD:AAY1028 BIFB133-13 MZB-BIF00158
6 BIF0498 West Java Kab Purwakarta Ci Asem -6.55703 107.675 BOLD:ACP6117 BIFD059-13 MZB-BIF00498