IDENTIFICATION OF PHEROMONE BINDING PROTEIN
GENE OF YELLOW RICE STEM BORER
Scirpophaga incertulas
(Walker) (Lepidoptera: Crambidae)
JAZIROTUL FITRIYATI
GRADUATE SCHOOL
BOGOR AGRICULTURAL UNIVERSITY
BOGOR
DECLARATION
I declare that this thesis entitled “Identification of Pheromone Binding Protein Gene of Yellow Rice Stem Borer Scirpophaga Incertulas (Walker)
(Lepidoptera: Crambidae)” was entirely completed by myself with resourceful help from the Department of Biology, Bogor Agricultural University. Information and quotes which were sourced from journals and books have been acknowledged and mentioned where in the thesis they appear. All complete references are given at the end of the paper.
Bogor, August 2011
Jazirotul Fitriyati
ABSTRACT
JAZIROTUL FITRIYATI. G352090151. Identification of Pheromone Binding Protein Gene of Yellow Rice Stem Borer Scirpophaga Incertulas (Walker) (Lepidoptera: Crambidae) (Supervised by RIKA RAFFIUDIN and I MADE SAMUDRA)
The yellow rice stem borers (YRSB) moth, Scirpophaga incertulas (Walker) (Lepidoptera: Crambidae), is known as the most major rice stem borer in tropical Asia. Pesticides are not effective to control the population of these insects due to almost entire larvae phase and pupae are in the rice stem. Hence, other control technique is needed such as based on the mating behaviour. Pheromone binding protein (PBP) in male S. incertulas antennae plays a role in the recognition of sex pheromone produced by the female, therefore influenced in their mating behavior. The aim of this study was to identify PBP gene of S. incertulas. For the purpose to collect imagoes as DNA source, S. incertulas were reared. Touchdown PCR and touchdown-nested PCR were the main techniques conducted to identify genomic of PBP gene from S. incertulas and revealed 700 and 600 bp amplicons, respectively. Those amplicons strongly expected as PBP gene. Sequence analysis of S. incertulas from touchdown-nested amplicon identified 575 bp which was consisted of 169 bp of exon 3 and 406 bp of intron 2. This study revealed putative amino acid sequences of exon 3 from S. incertulas has one conserved cysteine while other Lepidopterans PBP have three conserved cysteine. In phylogenetic analysis, the putative amino acid sequences obtained, showed a phylogenetic signal i.e. by clustering with PBPs from other Crambidae moths. The result of this study is important as a basic data for PBP expression analysis in female or male S. incertulas as the initial step to develope new insect biocontrol.
ABSTRAK
JAZIROTUL FITRIYATI. G352090151. Identifikasi Gen Pheromone Binding protein pada Penggerek Batang Padi Kuning Scirpophaga incertulas
(Walker) (Lepidoptera: Crambidae). (Dibimbing oleh RIKA RAFFIUDIN and I MADE SAMUDRA)
Ngengat Penggerek Batang Padi Kuning (PBPK) Scirpophaga incertulas (Walker) (Lepidoptera: Crambidae), diketahui sebagai hama penggerek batang padi utama di kawasan Asia tropis. Penggunaan pestisida tidak efektif untuk mengendalikan populasi serangga ini karena hampir seluruh fase larva dan pupa serangga ini berada di dalam batang. Oleh karena itu, teknik pengendalian lain diperlukan dalam pengendalian PBPK, seperti teknik pengendalian berdasarkan perilaku kawin. Pheromone binding protein (PBP) pada antena S. incertulas jantan mempunyai peranan dalam mengenali feromon sex yang dihasilkan oleh betina sehingga mempengaruhi perilaku kawin mereka. Tujuan dari penelitian ini adalah untuk mengidentifikasi gen PBP pada S. incertulas. Dilakukan pengembangbiakkan S. incertulas untuk mendapatkan imago sebagai sumber DNA. Touchdown PCR dan touchdown-nested PCR merupakan teknik amplifikasi utama yang dilakukan untuk mengidentifikasi data genom dari gen PBP pada S. incertulas dan menghasilkan dua amplikon berukuran 700 dan 600 pb berturut-turut. Kedua amplikon tersebut diduga kuat sebagai target dari gen PBP. Analisis sekuen dari S. incertulas dari amplikon hasil touchdown-nested dihasilkan sekuen berukuran 575 pb yang terdiri atas bagian ekson 3 (169 pb) dan intron 2 (406 pb). Penelitian ini menemukan sekuen asam amino putatif bagian ekson 3 PBP dari S. incertulas hanya memiliki satu sistein konservatif sedangkan PBP dari Lepidoptera lain memiliki tiga sistein konservatif. Dalam analisis filogenetik, sekuen asam amino putatif yang dihasilkan menunjukkan adannya sinyal filogenetik dengan mengelompok bersama PBP dari ngengat Famili Crambidae. Hasil penelitian ini sangat penting untuk digunakan sebagai data dasar untuk mempelajari analisis ekspresi PBP pada S. incertulas jantan dan betina sebagai langkah awal mengembangkan teknik baru pengendalian serangga hama.
SUMMARY
JAZIROTUL FITRIYATI. G352090151. Identification of Pheromone Binding Protein Gene of Yellow Rice Stem Borer Scirpophaga Incertulas (Walker) (Lepidoptera: Crambidae) (Supervised by RIKA RAFFIUDIN and I MADE SAMUDRA)
The yellow rice stem borers (YRSB) moth, Scirpophaga incertulas (Walker) (Lepidoptera: Crambidae), is known as the most major rice stem borer pest in tropical Asia. In China, India, and Southeast Asia, annual losses to yellow rice stem borers average 5 to 10%, but losses in individual fields may reach 50-60%. In Karawang, Indonesia, S. incertulas is dominant pest among other rice stem borers, and it caused 20,5% losses.
S. incertulas is monophagous in rice, its larva bore into the rice stem and complete the larva and pupa stages within the rice stem. During development, larva feed voraciously causing tremendous yield losses. Larva of S. incertulas are difficult to be controlled with insecticides because after hatching, the larva are exposed only for a few hours before they enter the rice stem. In addition, the use of insecticides can contaminate rice field and diminish the other non-target insect such as pollinator and predator.
Currently, Sustainable biocontrol such as mating disruption has implemented to control the population of S. Incertulas moth. Artificial sex pheromone trap as a famous example of mating disruption that using the analogue of sex pheromone which is produce by female moth to attract the male come to the trap. This method can lower the copulation behavior hence the pest population will decrease. The idea of artificial sex pheromone trap is influenced by natural mating behaviour of moth that using sex pheromone as communication signal. Female moth produce an airbone signal consisting of a single chemical compounds or a particular blend of compounds called sex pheromone. Moreover, male moth has pheromone binding protein (PBP) that selectively binds the sex pheromone release from the female moth for a signal to find the female position to mate.
PBP play a role in the recognition of sex pheromone in male moth antennae. By binding selectively to different components of pheromone blends, therefore, PBP can be a selective filter to bind the sex pheromone. Identification of PBP gene in S. incertulas is needed as an initial step to developing new insect biocontrol. Based on other species of Lepidoptera, PBP gene have two introns and three coding regions (exons); however, none was reported from S. incertulas.
incertulas was extracted using a Phenol/Chloroform method and amplification of PBP gene in S. incertulas was generated using eight degenerate primers. Five polymerase chain reactions (PCR) methods were used to amplify this gene and to improve the quality of generated PCR product, i.e. (i) Standard PCR, (ii) polyacrilamide gel cutting – re-PCR, (iii) nested PCR, (iv) touchdown PCR, and (v) touchdown-nested PCR. DNA sequencing and Phylogenetic analysis were done to identify PBP gene of S. incertulas.
Fifty seven S. incertulas imagoes emerged from the first rearing (parental collection from Bogor), seven male imagoes and 50 female imagoes. Whereas second rearing (parental collection from Karawang) emerged more number of imagoes than the first rearing, i.e. 88 imagoes consisted of 47 males and 41 females. Rain exposure was predicted as key factor that increased larval mortality rates during larva inoculation, hence emerged less imagoes than the second one. This is due to the first rearing was conducted in greenhouse covered with net roof; hence all first rearing process was exposed by rain. The second rearing was conducted in greenhouse with fiber roof, hence unexposed with rain. Life cycle span of S. incertulas from the first rearing was longer (48-56 days) than second rearing (44-51 days).
Although degenerate primers were designed in conserved region in Lepidopteran PBP genes, multi amplicons using standard PCR was produced. Polyacrilamide gel cutting-rePCR method revealed a single fragment from multi amplicons; however, those did not show the PBP features based on BLASTN (http://blast.ncbi.nlm.nih.gov/Blast.cgi) analysis hence, the cut amplicon was not the PBP target. Nested PCR still could not reduce non target fragments represented by multi amplicons having same pattern as amplicon with the same primers in standard PCR. This might be due to amplicon from standard PCR as DNA template in nested PCR, has several unselected fragments and caused misleading amplification when it was used in nested PCR as a DNA template. The last two PCR techniques touchdown PCR and touchdown-nested PCR gave one dominant fragment strongly expected as PBP target with 700 bp and 600 bp sizes, respectively.
gene, Odorant Binding Proteins (OBPs). The characteristic of PBP amino acid sequene are presence of six conserved cysteine residues, and so does the GOBP; therefore, BLASTP result obtained homology with GOBP. Might be due to A. mellifera is the second most complete genome data of insect; therefore, characterization of novel genomic data in insect will always hit to A. mellifera.
Alignment of the third exon putative amino acid sequencess PBPs among species showed there were three conserved cysteine residues, however, Since_PBPexpected amino acid only showed one concerved cysteine. Since_PBPexpected can be classified into PBP group by phylogenetic analysis and confirmed have a closest relationship with other Crambidae’s PBP (Ostrinia furnacalis and Ostrinia nubilalis). It has nearest distance with PBP of O. furnacalis (distance = 3.366) and PBP of O. nubilalis (distance = 3,366). Three disulfide bridges and hydrophobic pocket in PBP structure are formed by six conserved cysteine. Out of six conserved cysteine in PBP, three are located in exon 3 hence this region is more conserved than the other exons in PBP. Accordingly, even only one conserved cysteine was found in third exon of Since_PBPexpected, this sequence still gave phylogenetic signal by clustering with other PBPs of Crambidae.
RINGKASAN
JAZIROTUL FITRIYATI. G352090151. Identifikasi Gen Pheromone Binding protein pada Penggerek Batang Padi Kuning Scirpophaga incertulas
(Walker) (Lepidoptera: Crambidae). (Dibimbing oleh RIKA RAFFIUDIN dan I MADE SAMUDRA)
Ngengat penggerek batang padi kuning (PBPK), Scirpophaga incertulas (Walker) (Lepidoptera: Crambidae), diketahui sebagai hama penggerek batang padi yang utama di kawasan Asia tropis. Di Cina, India, dan Asia Tenggara, persentase kehilangan hasil panen karena PBPK rata-rata 5-10% per tahun, namun kehilangan hasil panen karena PBPK pada sawah individual dapat mencapai 50-60%. Di Karawang, Indonesia, S. incertulas merupakan hama penggerek batang padi yang paling dominan dibandingkan dengan hama penggerek lainnya, dan kerugian yang ditimbulkan akibat hama ini mencapai 20,5%.
S. incertulas merupakan serangga monofag pada padi, larva dari S. incertulas akan menggerek ke dalam batang padi dan menyelesaikan fase larva dan pupa di dalam batang padi. Selama masa perkembangan, larva memakan batang padi dengan sangat rakus sehingga dapat menimbulkan kehilangan produksi gabah yang signifikan. Larva dari S. incertulas sulit dikendalikan menggunakan insektisida karena segera setelah menetas, larva hanya akan terekspose beberapa jam di luar lalu akan segera memasuki batang. Selain itu, penggunaan insektisida secara berlebihan dapat menimbulkan terjadinya kontaminasi pada sawah dan dapat membunuh serangga-serangga lain yang menguntungkan seperti predator atau musuh alaminya.
Saat ini, penggunaan teknik pengendalian hama yang berkelanjutan seperti Penggangguan proses kawin telah dilakukan untuk mengendalikan populasi ngengat S. Incertulas. Perangkap feromon sex buatan merupaka salah satu metode penggangu proses kawin yang paling dikenal. Perangkap ini menggunakan analog dari feromon sex yang dihasilkan oleh ngengat betina untuk menarik ngengat jantan untuk mendatangi perangkap. Metode pengendalian ini dapat menurunkan perilaku kawin sehingga dapat menurunkan populasi hama generasi berikutnya. Gagasan pembuatan perangkap ini di dasari dari perilaku kawin ngengat secara alami di alam yang menggunakan feromon sex sebagai sinyal komunikasi. Ngengat betina menghasilkan sinyal mudah menguap yang terdiri dari senyawa kimia tunggal atau campuran dari senyawa tertentu yang disebut dengan feromon sex. Sedangkan pada ngengat jantan memiliki pheromone binding protein (PBP) yang akan mengikat secara selektif feromon sex yang dihasilkan ngengat betina sebagai sinyal untuk menemukan posisi betina lalu melakukan kopulasi (kawin).
beberapa spesies dalam Lepidoptera, gen PBP memiliki dua intron dan tiga exon, namun belum terdapat data tentang gen PBP pada S. incertulas.
Imago S. incertulas jantan dan betina dikembangbiakkan sebanyak dua kali dengan tujuan untuk mendapatkan imago sebagai sumber DNA. Pengembangbiakkan S. incertulas yang pertama dilaksanakan pada tanggal 6 September – 26 Oktober 2010 dan pengembangbiakkan yang kedua dilaksanakan pada tanggal 4 Oktober – 15 November 2010. Pengembangbiakkan pertama menggunakan imago indukan yang berasal dari Bogor sedangkan yang kedua menggunakan imago indukan yang berasal dari Karawang. Proses pengembangbiakkan S. incertulas dilaksanakan dalam rumah kaca. Selanjutnya DNA total dari S. incertulas diekstraksi menggunakan metode fenol/kloroform dan Amplifikasi gen PBP pada S. incertulas dilakukan dengan menggunakan primer degenerate. Sebanyak lima metode polymerase chain reaction (PCR) dilakukan untuk meningkatkan kualitas produk PCR (amplikon) yang dihasilkan, yaitu: (i) standard PCR , (ii) polyacrilamide gel cutting-rePCR, (iii) nested PCR, (iv) touchdown PCR, (v) touchdown-nested PCR.
Sebanyak 57 imago dihasilkan dari proses pengembangbiakkan yang pertama dengan rincian yaitu tujuh imago jantan dan 50 imago betina. Sedangkan hasil pengembanbiakkan S. incertulas yang kedua didapatkan 88 imago dengan rincian 47 jantan dan 41 betina. Paparan hujan diduga sebagai factor kunci yang meningkatkan laju mortalitas larva pada saat inokulasi larva, sehingga pada pengembangbiakkan yang pertama dihasilkan jumlah imago yang lebih sedikit dibandingkan dengan yang kedua. Hal ini disebabkan karena pengembangbiakkan tahap pertama dilakukan pada rumah kaca yang atapnya terbuat dari jaring sehingga air hujan dapat masuk ke dalam. Pengembangbiakkan S. incertulas yang kedua dilakukan pada rumah kaca yang atapnya terbuat dari fiber sehingga air hujan tidak dapat masuk ke dalam rumah kaca. Lama siklus hidup S. incertulas dari pengembangbiakkan tahap pertama lebih lama (48-56 hari) dibandingkan pengembangbiakkan tahap kedua (44-51 hari).
diduga kuat sebagai amplikon target gen PBP yaitu touchdown PCR (700 pb) dan touchdown-nested PCR (600 pb).
Terdapat enam sekuen yang dihasilkan dari enam amplikon yan menggunakan tiga metode PCR yang berbeda: (i) metode polyacrilamide gel cutting-rePCR (dua sekuen: 217 pb (primer F2.1-R3.1) dan 235 bp (primers F2.2-R3.1), (ii) nested PCR (tiga sekuen: 265 pb, 317 pb, dan satu sekuen dari amplikon berukuran 700 pb tidak dapat terbaca), (iii) touchdown-nested PCR (satu sekuen: 575 bp). Pada 4 hasil sekuen dari amplikon metode polyacrilamide gel cutting-rePCR dan nested PCR tidak ditemukan adanya Open Reading Frame (ORF). Hasil BLASTN dari keempat sekuen tersebut juga tidak menunjukkan adanya karakteristik gen PBP pada sekuen tersebut. Amplikon dari metode touchdown-nested PCR dapat menghasilkan sekuen sepanjang 575 pb dari S. incertulas (dinamakan Since_PBPexpected) yang terdiri dari ORF sepanjang 169 pb (56 asam amino) dan bagian intron sepanjang 406 pb. Bagian ORF sekuen tersebut diduga sebagai bagian dari ekson 3 dari gen PBP, sedangkan bagian intron yang teridentifikasi diduga sebagai bagian intron 2 dari gen PBP pada S. incertulas. Ujung 3’ pada bagian intron dari sekuen ini memiliki struktur AG yang menunjukkan batas antara intron dan ekson. Hasil BLASTP (http://blast.ncbi.nlm.nih.gov/Blast.cgi) dari 56 asam amino hasil translasi dari ORF pada sekuen ini menujukkan bahwa sekuen asam amino tersebut memiliki homologi dengan gen General Odorant Binding Protein (GOBP) pada Apis mellifera. GOBP dan OBP merupakan anggota dari family gen Odorant Binding Protein (OBP). Karakteristik adanya enam asam amino sistein konservatif yang terdapat pada PBP juga terdapat pada GOBP. Karakteristik yang dimiliki bersama oleh PBP dan GOBP ini diduga sebagai penyebab hasil BLASTP dari sekuen asam amino Since_PBPexpected memiliki homologi dengan GOBP. Selain itu, A. mellifera merupakan serangga dengan data genom terlengkap kedua di Genbank, hal ini yang menyebabkan karakterisasi data genom baru dari serangga akan sering mengacu pada A. mellifera.
© Copyright of IPB, year 2010
Copyright reserved
1. Forbidden to quote part or all of these writings without including or mentioning the source.
a. Be cited only for educational purposes, research, writing papers, drafting reports, writing criticism or review an issue;
b. Quotation must not harm the affairs of IPB.
IDENTIFICATION OF PHEROMONE BINDING PROTEIN
GENE OF YELLOW RICE STEM BORER
Scirpophaga incertulas
(Walker) (Lepidoptera: Crambidae)
JAZIROTUL FITRIYATI
A Thesis
As Partial fulfillment of the Requirement to obtain Master of Science Degree in Animal Biosciences
GRADUATE SCHOOL
BOGOR AGRICULTURAL UNIVERSITY
BOGOR
External Member Supervisor Committee:
Title : Identification of Pheromone Binding Protein Gene of Yellow Rice Stem Borer Scirpophaga incertulas (Walker) (Lepidoptera:Crambidae)
Name : Jazirotul Fitriyati Registration Number : G352090151 Major : Animal Biosciences
Approved:
Advisory Committee
Dr. Ir. Rika Raffiudin, M.Si Dr. Ir. I Made Samudra, M.Sc (Chair) (Member)
Agreed:
Coordinator of Animal Biosciences Major Dean of Graduate School
Dr. Bambang Suryobroto Dr. Ir. Dahrul Syah, M.Sc. Agr
ACKNOWLEDGEMENT
This research was financially supported by Kerja Sama Kemitraan Penelitian Pertanian Dengan Perguruan Tinggi (KKP3T) from Ministry of Agriculture Indonesia Republic and Bakrie Center Foundation (BCF) for Masters Scholarship funding.
I would like to express my gratitude for Dr. Ir. Rika Raffiudin, M.Si and Dr. Ir. I Made Samudra, M.Sc as supervisory committee, for all of guidance and encouragement as well as invaluable academic advices for the whole period of my study and research at IPB.
I am very grateful to all the invaluable lecturing staff (Especially for Dr. Achmad Farajallah) and technical staff of Animal Biosciences Study Program who have imparted knowledge and help and also for the full support given to enable the successful completion of this research
Moreover, I would like to express my appreciation to all fellow students of Animal Bioscience especially for class of 2009. Of these fellows, I would like to express my deepest thanks to close friends Ruth Martha Winnie, Ibu Taruni Sri Prawasti, Sanou Faye, Kaleem Saleem, Petlane Molafe, and Princy Razafimandimby who always providing a helping hand and good advices. I wholeheartedly thank to research team of Dr. Rika Raffiudin and Dr. Achmad Farajallah supervisees such as Cahyo, Made, Rindi, Raisa, Bisri, Dea, Rijal, and Chyntia who made my master research a great experience of team working.
Further, I am highly indebted to my affectionate parents, young brothers (Reza, Kiki, and Agil) and other family members who always inspire and encourage me for higher education, and finally to Yonan Arman who contributes immensely in providing supports and good constructed advice during busy time of my study and research.
BIOGRAPHY
Jazirotul Fitriyati was born on January 5th, 1988 in Kendal, Central Java as the first daughter of Mr Jazuri and Mrs Sri Nuriyati among other three children.
In 2005, Ms fitriyati finished Senior High School from State Senior High School 6 Semarang, she continued to study for Bachelors Degree in Department of Biology, Mathematic and Natural Sciences Faculty, Bogor Agricultural University and graduated in June 2009. Ms Fitriyati was awarded by the Bakrie Center Foundation to continue her study to Masters Degree in Bogor Agricultural University, majoring in Animal Biosciences in 2009.
TABLE OF CONTENTS
Page
Standard PCR ... 22 Polyacrilamide gel cutting – RePCR method ... 22 Nested PCR ... 23 Touchdown PCR ... 24 Touchdown-nested PCR ... 24 Identification of the PBP Expected Sequences from S. incertulas ... 25 Phylogenetic Analysis of Third Exon ... 27 PBP Sequence of S. incertulas ... 27 5. DISCUSSION ... 31
Rain exposure as a key factor that influenced the emergence of S. incertulas imago ... 31 Optimalization amplicon of S. incertulas PBP gene by using degenerate
primers and touchdown-nested PCR method ... 32 Clustering Since_PBPexpected to the PBP of Crambidae based on
Phylogenetic Analysis ... 34 Future Research ... 35 6. CONCLUSION AND SUGGESTION ... 37 Conclusion ... 37 Suggestion ... 37 REFERENCES ... 38 APPENDICES ... 43
LIST OF TABLES
Page
1 Sex pheromone compounds ratio from several moths with different functional group ... 7 2 Several pheromone binding proteins gene data of Lepidoptera member in
Genbank ... 10 3 PCR and sequencing degenerate primers for PBP Gene of Scirpophaga
incertulas ... 16 4 Primer combinations list that were used to amplify PBP gene of Scirpophaga
incertulas ... 17 5 List of PBP, OBP, GOBP, and OR sequences used for phylogenetic analysis. 20 6 Life cycle of reared Scirpophaga incertulas ... 21 7 Top one BLASTN result of two fragments sequenced from Scirpophaga
incertulas polyacrilamide gel cutting rePCR amplicon ... 26 8 Top one BLASTN result of two fragments sequenced from Scirpophaga
incertulas nested PCR amplicon ... 26 9 BLASTP result from third exon putative amino acid sequencess of Scirpophaga incertulas touchdown-nested PCR amplicon ... 27 10 Genetic distance of Scirpophaga incertulas PBP putative amino acid
LIST OF FIGURES
Page
1 Praecinctorium: absent in Pyralidae (A & B) and present in Crambidae ... 3 2 Life cycle of the yellow rice stem borer (YRSB) Scirpophaga incertulas ... 4 3Diagrammatic section through a sensillum of Lepidoptera ... 8 4 Diagrammatic of the binding protein capturing pheromone molecules in lymph
sensilla ... 8 5 Binding of two different pheromones to the Antheraea polyphemus PBP
binding site ... 9 7 Equipments to capture imago of Scirpophaga incertulas in the field ... 12 8 Geenhouse condition was used for Scirpophaga incertulas rearing process .... 12 9 Rearing method of rice stem borer ... 13 10 Position of eight degenerate primers in alignment of five PBP sequences ... 15 11 Primer position used to amplify Scirpophaga incertulas based on genomic
PBP gene of Ostrinia furnacalis ... 16 12 Expected PBP amplicons of Scirpophaga incertulas using twelve primer
combinations ... 22 13 Expected PBP amplicons of Scirpophaga incertulas using polyacrilamide gel
cutting – RePCR modification ... 23 14 Expected PBP amplicons of Scirpophaga incertulas using nested PCR
modification ... 23 15Expected PBP amplicon of Scirpophaga incertulas using touchdown PCR
modification ... 24 16 Expected PBP amplicons of Scirpophaga incertulas using touchdown- nested
PCR modification ... 25 17 DNA sequence and predicted amino acid sequence of Scirpophagaincertulas
PBP expected from Touchdown-nested amplicon ... 26 18 BLASTP alignment result of Since_PBPexpected amino acids sequence with
19 Alignment of the amino acid sequences from translated third exon of
Scirpophaga incertulas PBP expected. ... 28 20 Conserved cysteine amino acid position in exons of three moths PBP gene and
the schematic of intron and exon size from three moths PBP gene ... 28 21 Phylogenetic analysis of Since_PBPexpected PBP with other OBP, GOBP,
PBP, and OR genes of Crambidae moths based on amino acid sequences using Maximum likelihood method implemented WAG model, with 1000 Bootstrap 1000 replication. ... 29 22Schematic diagram of the three disulfide linkages in PBP of Bombyx mori .. 35
LIST OF APPENDICES
Page
1 Chromatogram of Scirpophaga incertulas polyacrilamide gel cutting – RePCR amplicon using pair of sequencing primer F2.1 and R3.1 ... 44 2 Chromatogram of Scirpophaga incertulas polyacrilamide gel cutting – RePCR
amplicon using a pair of sequencing primer F2.2 and R3.1 ... 45 3 Chromatogram of Scirpophaga incertulas nested PCR amplicon from 700 bp
fragment using reverse primer R3.1 ... 46 4 Chromatogram of Scirpophaga incertulas nested PCR amplicon from 500 bp
fragment using reverse primer R3.1 ... 47 5 Chromatogram of Scirpophaga incertulas nested PCR amplicon from 300 bp
fragment using forward primer F2.1 ... 48 6 Chromatogram of Scirpophaga incertulas touchdown-nested PCR amplicon
1. INTRODUCTION
Background
The yellow rice stem borers (YRSB) moth, Scirpophaga incertulas (Walker) (Lepidoptera: Crambidae), is known as the most major rice stem borer pest in tropical Asia (Kumar et al. 2001). In China, India, and Southeast Asia, annual losses to yellow rice stem borers average 5 to 10%, but losses in individual fields may reach 50-60% (Taylor 1988). In Karawang, Indonesia, S. incertulas is dominant pest among other rice stem borers, and it caused 20,5% losses (Suharto and Usyati 2005).
S. incertulas is monophagous in rice (Pathak and Khan 1994), its larva bore into the rice stem and complete the larva and pupa stages within the rice stem. During development, larva feed voraciously causing tremendous yield losses (Kumar et al. 2001). Larva of S. incertulas are difficult to be controlled with insecticides because after hatching, the larva are exposed only for a few hours before they enter the rice stem. In addition, the use of insecticides can contaminate rice field and diminish the other non-target insect such as pollinator and predator (Pathak and Khan 1994, Cork and Hall 1998).
of heksadecenyl aldehyde, cis-11-heksadecenyl aldehyde, and cis-9-octadecynyl aldehyde (Tatsuki et al. 1985).
PBP gene consists of two introns and three coding regions (exons) (de Santis et al. 2006; Xiu and Dong 2007; Xiu et al. 2008), as shown in PBP gene of Crambidae Family i.e Chilo supressalis (Striped rice stem borer) (Genbank EU825762), Ostrinia furnacalis (Asian corn stem borer) (Genbank AF133629) and Ostrinia nubilalis (European corn borer) (Genbank AF133643). PBP plays a role in the recognition of sex pheromone in male moth antennae. By binding selectively to different components of pheromone blends, therefore, PBP can be a selective filter to bind the sex pheromone (Willett and Harrison 1999). Out of all the PBP gene found in Lepidoptera, none was reported from S. incertulas. Identification of S. incertulas PBP gene can be used as an initial step to determine protein structure and to developing new insect control using the PBP inhibitor to decrease capability by the insect to detect new incoming pheromone; hence, prevent them to mate (Riba et al. 2001).
Research Objective
These researches were aimed to identify and characterize the genome of PBP gene in S. incertulas.
Research Output
Pheromone Binding Protein genomic data of S. incertulas will be used as basic information for studying PBP expression in female or male S. incertulas; moreover, for initial step to developing new insect biocontrol.
Hypotheses
This current study proposed several hypotheses are:
1. Pheromone binding protein gene of S. incertulas might have high similarity with Crambidae’s PBP gene due to their close relationship.
2. Intron and exon position on O. furnacalis PBP gene is suppose to be similar with that of S. incertulas PBP gene.
2. LITERATURE REVIEW
Classification and Distribution of Scirpophaga incertulas
Scirpophaga incertulas is classified in Order of Lepidoptera, Family of
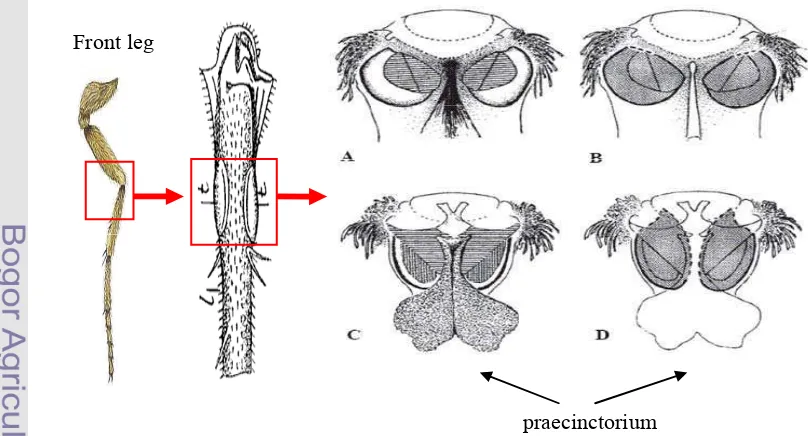
Crambidae, and Subfamily of Schoenobiinae (Kristensen 2007). Crambidae has 17 subfamilies and more than 11.600 species known today (Solis 2007). Many Crambidae moth involved as pest and usually called the grass moth family. Chilo and Ostrinia are the best known genus for biology and genetics data in Crambidae. Previously, Crambidae have been treated as a subfamily of the Pyralidae or snout-moths, but currently, Crambidae is separated from Pyralidae. The principal difference between Crambidae and Pyralidae are the structure in the tympanic at the front leg called the praecinctorium, which joins two tympanic membranes in the Crambidae; on the contrary, it is absent in the Pyralidae (Figure 1).(Solis 2007).
[image:33.595.99.503.411.629.2]Distribution of S. incertulas is throughout of Asia such as Afghanistan, Pakistan, India, Nepal, Burma, China, Thailand, Vietnam, Cambodia, Malaysia, Indonesia, Sabah, Philippines, Hongkong, Taiwan, Japan, and Okinawa (Grist and
Figure 1 Praecinctorium: absent in Pyralidae (A & B) and present in Crambidae (C & D) (Solis 2007)
Front leg
Lever 1969, Pathak and Khan 1994). Chilo distribution was recorded as a pest in Portugal, Spain, Iraq, Pakistan, India, Sri Langka, Vietnam, Cambodia, Laos, Malaysia, Indonesia, Thailand, Sabah, Philippines, New Guinea, Taiwan, Korea, China, and Japan (Grist and Lever 1969).
Morphological Structure and Life Cycle of S. incertulas
Scirpophaga incertulas undergoes metamorphosis from egg, larva, pupae, up to adult. The eggs hatch in 5 to 9 days, and the optimum egg hatching temperature is 24-29 °C. After hatching, the first instar larva feed on leaf tissue before bore the rice stem. Larva of S. incertulas usually undergoes four to seven larval instars stages to become full grown larva. The larval development take 20 to 30 days, followed by a pupal period of 9-12 days. The average imago life is only 5 to 7 days for mating including oviposit their eggs. The entire life cycle of S. incertulas range from 39 to 58 days (Figure 2). (Pathak and Khan 1994).
YRSB Adult
5‐9 days
20‐30 days 9‐12 days
Figure 2. Life cycle of the yellow rice stem borer (YSB) Scirpophaga incertulas (Pathak and Khan 1994)
Figure 2 Life cycle of the yellow rice stem borer (YRSB) Scirpophaga incertulas (Pathak and Khan 1994) Egg mass
5-9 days
4-7 Larval stages
20-30 days
Larva Pupa
9-12 days
The eggs of S. incertulas are laid in a flat compact mass covered with brown scales measuring 0.14 x 0.11 mm length; color translucent at first, darkening during development. An egg mass contain 50 to as many as 150, with an average total of 600 eggs per female. The larva of S. incertulas is ivory-colored to greenish and rather worm-like in appearance, posterior tapering from the first abdominal segment. There are four instars; length 18 to 22 mm. The pupa of S. incertulas have a straw colored within a white silken cocoon measuring 18 to 20 mm, the pupa itself being 13 to 15 mm long. The adult male has a forewing brownish ochre with a rather indistinct black spot at the lower angle of the cell, an opaque brown band and marginal row of black dots, hindwings white, and span 18 to 23 mm. The adult female has an orange-yellow forewing with a prominent black spot, hindwing light fuscous, and span 24 to 36 mm in female (Grist and Lever 1969, Pathak and Khan 1994).
The Damages Are Caused by S. incertulas
Being the most important insect pest in the world, S. incertulas is responsible for destroying an average of about 0.8 million tons of rice a year, while crop losses in Taiwan vary from 20 to 40 percent (Grist and Lever 1969). In Indonesia, the infestation intensity of rice borer and the damage area in 1988 were 20.5% and 151.577 ha, respectively (Suharto and Usyati 2005). Crop losses which are caused by yellow rice stem borer occurred in China, India, and Southeast Asia, annual losses to rice borer average 5 to 10%. Recovery or prevention of 5% of the losses to stem borers could feed 140 million people for 1 year (Taylor 1988).
panicles are whitish and unfilled or empty (Pathak and Khan, 1994; Cork and Hall 1998).
Yellow rice stem borer are difficult to control with insecticides because after hatching, the first instar larva are exposed only for a few hours before they enter a tiller or enter other part of the rice stem. In addition, the use of insecticides has side effect such as water contamination and indiscriminate killing (Pathak and Khan 1994; Cork and Hall 1998). Currently, Sustainable biocontrol such as mating disruption has implemented to control the population of S. Incertulas moth. Artificial sex pheromone trap as an example of mating disruption that using the analogue of sex pheromone which is produce by female moth to attract the male come to the trap. This method can lower the copulation behavior hence the pest population will decrease (Cork and Hall 1998).
The idea of artificial sex pheromone trap is influenced by natural mating behaviour of moth that using sex pheromone as communication signal. Female moth is release sex pheromone into the air to attract male moth (Reelofs 1995). Moreover, male moth has pheromone binding protein (PBP) that selectively bind the sex pheromone release from the female moth for a signal to find the female position to mate (Willett and Harrison 1999). In the same case, male moth will be attracted to artificial pheromone as a presence of female insect and they will be able to respond by flying to the trap (Willett and Harrison 1999; Tatsuki et al. 1985).
Sex Pheromone and PBP Proteins Function in Moth Mating Behavior
Female moth produce an airbone signal consisting of a single chemical compounds or a particular blend of compounds called sex pheromone (Reelofs 1995). A typical Lepidoptera or moth’s sex pheromone consists of two or three structurally related compounds in a specific ratio. The compounds often contain an oxygenated functional group that function as terminal groups such as acetates, alcohols or aldehydes (Cork et al. 2007) (Table 1).
in the male moth’s antennae (Willet and Harrison 1999). Pheromone binding protein was first identified by its ability to bind the sex pheromone of the silk moth Antheraea polyphemus (Vogt and Riddiford 1981).
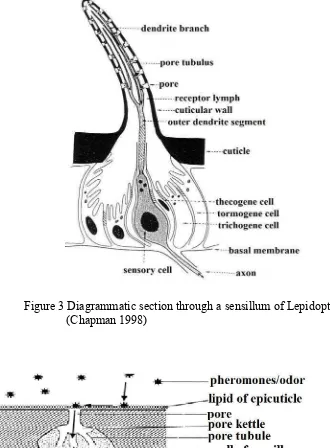
Sex pheromone and their receptors, PBP are the important component which playing the role of mating behavior in insect, especially in moths. Most of pheromone molecules are lipophilic, so they will dissolve in the epicuticular lipid forming the outer coating of a sensillum on the male antennae, and then they will enter into the lymph through pore and pore tubule (Figure 3). Pheromone molecules which entered the lymph will be captured by PBP receptor and transported to receptor molecules in the dendrite membrane as a signal of female moth presence (Figure 4) (Merrit et al. 1998). Male moth will respond these signal, they are flying upwind and finding the female for mating (Roelofs et al. 1995). The antennae of male moths are extremely sensitive to the female sex pheromone such that a male moth can find a female up to 4.5 km away (Vogt and Riddiford 1981). S. incertulas is a nocturnal insect, so they do mating and oviposition in the night (Pathak and Khan 1994).
Table 1 Sex pheromone compounds ratio from several moths with different functional group
No Species Family
Sex pheromone compounds Ratio (%) Functional
group Reference
1 S. incertulas Crambidae
E9-16: Ald 17 Aldehyde
Tatsuki et al.
(1985) E11-16: Ald 66 Aldehyde
E9-18: Ald 17 Aldehyde
2 O. furnacalis Crambidae E12-14:OAc 53 Acetate
Willet and Harrison (1999)
Z12-14:OAc 47 Acetate
3 O. nubilalis Crambidae E11-14:OAc 99 Acetate
Z11-14:OAc 1 Acetate
4 Grapholita molesta Tortricidae
Z8-12:OAc 95 Acetate
Han et al.(2001)
E8-12:OAc 4 Acetate
Figure 3Diagrammatic section through a sensillum of Lepidoptera (Chapman 1998)
The Structure of Pheromone Binding Protein
Pheromone Binding Protein and General Odorant Binding Protein (GOBP) are members of a larger gene family, the insect Odorant Binding Protein (OBPs), which are small, water-soluble protein expressed in the olfactory sensillum tissue of insects (Vogt et al.1991). Unlike GOBP that have a role in detection of general odorants from food and host plant, PBP only detect sex pheromone molecules to specific neural receptors in male antennae (Vogt et al. 1989; Prestwich et al. 1995). Initially defined by their ability to bind sex pheromone, PBP are found in very high concentrations (10 mM) in the sensillar lymph that surrounds dendrites of pheromone sensitive neurons within the olfactory sensilla along the antennae (Vogt and Riddiford 1981, Vogt et al. 1989). This protein of about the same size (15 kDa) (Pleosi and Maida 1995) and have six conserved cysteines residues in their amino acids that is cleaved off in the mature protein binding. Expression studies from this protein have led to a model of PBP function in which pheromone is bound into a hydrophobic pocket of the PBP (Figure 5). Hydrophobic pocket (HP) in PBP allowing the long chain hydrocarbons to cross the aqueous barrier of the sensillar lymph and interact with specific receptors such as Odorant Receptor (OR) on the surface of pheromone-sensitive neurons (Vogt et al. 1989; Prestwich et al. 1995).
In their role of binding and solubilizing pheromone, PBP may act as selective filters for the female pheromones by specifically binding and solubilizing the appropriate pheromone molecules. Pheromone binding protein genes have been described in some species of Lepidoptera from Family of Crambidae, Sphingidae, Noctuidae, Saturniidae, and Yponomeutidae (Table 2). Based on genomic data of PBP gene in O. furnacalis, it has two introns and three coding regions (exons) (Willett 2000; Willett and Harrison 1999) (Figure 6).
[image:40.595.85.505.268.742.2]
Figure 6 Schematic intron and exon location of Ostrinia furnacalis PBP gene (constructed based on Willett and Harrison (1999))
Table 2 Several pheromone binding proteins gene data of Lepidoptera member in Genbank
No Species Family
Length (bp)
Genbank Acc
number References
1 Manduca sexta Sphingidae 841 AF117588 Robertson et
al. (1999)
2 Lymantria dispar Noctuidae 672 AF007867 Merritt et al. (1998)
3 O. furnacalis Crambidae 1741 AF133629 Willett and Harrison (1999)
4 O. nubilalis Crambidae 1479 AF133643 Willett and Harrison (1999)
5 Antheraea polyphemus Saturniidae 1076 AJ277266 Maida et al. (2000)
6 Yponomeuta cagnagellus Yponomeutidae 2945 AF177661 Willett (2000)
3. MATERIAL AND METHOD
Time and Place of Research
This research was conducted from April 2010 up to May 2011. S. incertulas imagoes were collected from several areas in Bogor and Karawang (West Java). The stem borers were reared in green house of the Indonesian Centre for Agricultural Biotechnology and Genetic Resources Research and Development (BB Biogen). Molecular and data analysis was conducted in Division of Animal Biosciences, Department of Biology, Bogor Agricultural University (IPB).
Material
Male and female imagoes of S. incertulas that used in this study were collected from Bogor and Karawang and reared in the green house of BB Biogen. Collected and reared imagoes were preserved in absolute ethanol to preserve the DNA and samples were kept at refrigerator as collection of Molecular laboratory of Animal Biosciences, Department of Biology, Bogor Agricultural University (IPB).
S. incertulas Imagoes Collection
S. incertulas Imagoes Rearing
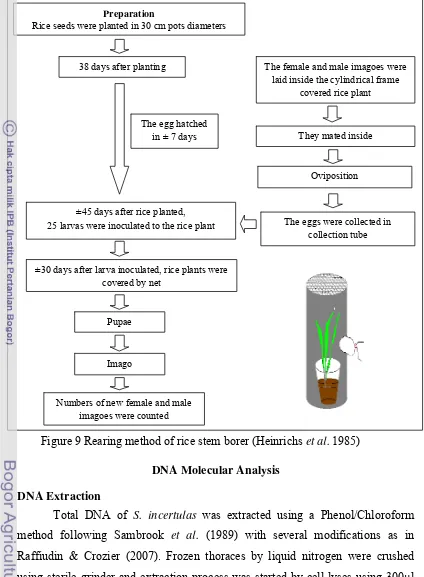
[image:42.595.328.421.81.191.2]Male and female imagoes of S. incertulas were reared two periods for the purpose to collect imagoes as DNA source, first during September 6th - October 26th 2010 and the second during October 4th – November 15th 2010 . The first rearing used male and female parental imagoes of S. incertulas that were collected from Bogor and the second used those from Karawang. Both rearing were conducted in a greenhouse which has two type of roofs i.e. net roof (rain exposed) (Figure 8b) for the first rearing and fiber roof (rain unexposed) for the second (Figure 8a). Fifty pots were used for each rearing for rice planting (Ciherang variant) were then rearing method of stem borer followed Heinrichs et al. (1985). Rearing steps consisted of several processes i.e. rice seedlings, rice planting, parental imagoes collection, larva inoculation, and the release of imago (Figure 9). Thirty days after larva inoculation, one or two rice stems that showed whiteheart symptoms caused by larval infestation were dissected to examine the pupa stage of S. incertulas. Each step of the life cycle (egg-imagoes) was recorded to estimate the life cycle of S. incertulas.
Figure 7Equipments to capture imago of Scirpophaga incertulas in the field
Insect net Collection tube
Figure 8 Geenhouse condition was used for Scirpophaga incertulas rearing process. A: Greanhouse was covered by fiber roof (rain unexposed), B: Greenhouse was covered by net roof (rain exposed)
DNA Molecular Analysis
DNA Extraction
Total DNA of S. incertulas was extracted using a Phenol/Chloroform method following Sambrook et al. (1989) with several modifications as in Raffiudin & Crozier (2007). Frozen thoraces by liquid nitrogen were crushed using sterile grinder and extraction process was started by cell lyses using 300µl 2% N-Cethyl-N,N,N-Trimethyl Ammonium Bromide (CTAB) and 10µl proteinase-K (5 mg/ml) digestion. The homogenate was incubated at 55˚C for 2 hours and gently mixed every 30 minutes in order to activate proteinase-K. The mixture was centrifuged for 10 minutes at 13000 rpm. The supernatant was transferred into a new tube for purification process. The DNA was extracted from
Preparation
Rice seeds were planted in 30 cm pots diameters
38 days after planting The female and male imagoes were laid inside the cylindrical frame
covered rice plant
They mated inside
Oviposition
The eggs were collected in collection tube The egg hatched
in ± 7 days
±45 days after rice planted, 25 larvas were inoculated to the rice plant
Pupae
Imago
Numbers of new female and male imagoes were counted
[image:43.595.96.520.63.640.2]±30 days after larva inoculated, rice plants were covered by net
it by adding two times an equal volume of using Phenol:Chloroform:Isoamylalcohol (PCI) (25:24:1) and followed by adding two times an equal volume of Chloroform:Isoamylalcohol (CIAA) (24:1). Each time of PCI and CIAA step, prior to the samples were centrifuged (13000 rpm; 5 min), those were mixed gently to collect the aqueous phase. The purified DNA was precipitated by adding 360µl Isopropanol which incubated at 4˚C overnight. The mixture was centrifuged at 13 000 rpm (30 min; 10˚C) and pellets were washed by adding 300µl 70% ethanol and subsequently centrifuged at 13000 rpm (10 min). Subsequently pellets were dried using vacuum machine to evaporate alcohol residual. Total DNA from dried pellets were diluted in 30µl TE (2M Tris-HCL: 0.2 M EDTA) buffer (pH 8.0) and stored in freezer.
DNA Amplification
Eight degenerate primers were designed to match conserved regions of the previously published PBP sequences from two PBPs identified of C. supressalis (Genbank GU321120, EU825762), one PBP from O. nubilalis (Genbank AF133643), one PBP from O. furnacalis (Genbank AF133629), and one PBP type 1 from Spodoptera exigua (Genbank AY540316) (Figure 10). There were two forward primers in exon one (PBP_Sc_F1.1 and PBP_Sc_F1.2), two forward primers in exon two (PBP_Sc_F2.1 and PBP_Sc_F2.2), four reverse primers in exon two (PBP_Sc_R2.1 and PBP_Sc_R2.2) and exon three (PBP_Sc_R3.1 and PBP_Sc_R3.2) (Figure 9). Degenerate primers sequences are listed in Table 3.
Multi band PCR products were produced by using degenerate primers, therefore, I developed several PCR modifications to improve PCR product. All PCR techniques were carried out in ESCO thermal cycler, and these were several PCR techniques performed as mentioned below:
Standard PCR
Amplification was performed in 50 µl volumes using 2X ReadyMix (Kapa, South Africa) containing reaction buffer with 1,5 mM Mg2+, 0.4 mM each dNTP, 0,4 µM each forward and reverse primer and 2.5 Unit Taq DNA polymerase and 10-100 ng DNA template. Polymerase chain reaction was conducted in 5 min at 940C for initial denaturing, 30 cycles of 1 minutes at 940C for denaturing, 1 minutes at 500C for annealing and 1,5 minutes at 720C for DNA elongation, followed by 5 minutes at 720C for the final extension.
Table 3 PCR and sequencing degenerate primers for PBP Gene of Scirpophaga incertulas
Name Primer sequence (5'-3') Position Target region
PBP_Sc_F1.1 GYGGARTSSTCKCAGGAKRTCATGA exon1 VESSQEIMK*
PBP_Sc_R2.1 TSASCKCATAGYCWTCCTTCCAG exon2 WKDDYAVT
PBP_Sc_F1.2 TTYGGSAAAGCKTWSGAYASSTG exon 1 FGKAYDSC
PBP_Sc_R2.2 AGCTTKKWGGASAKGCACATRAT exon 2 IMCMSNKL
PBP_Sc_F2.1 GAGTTCGCYAAGAAACATGG exon 2 REFAKKH*
PBP_Sc_R3.1 CTTGTGRATCTCSRCCTTGAAGCA exon 3 CFKVEIHK
PBP_Sc_R3.2 ATCAGGTCCATRCTGGGAGCCCA exon 3 WAPSMDLIM
PBP_Sc_F2.2 TGTGCMTSTCCWMMAAGCTSGACCT exon 2 CMSNKLDL
* Primers were in the same region as those designed by Abraham et al. (2005)
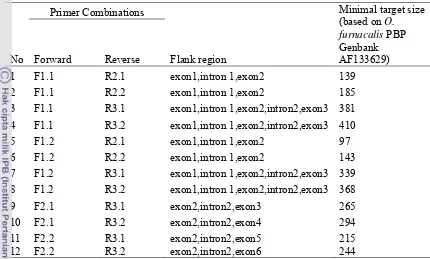
Table 4 Primer combinations list that were used to amplify PBP gene of Scirpophaga incertulas
No
Primer Combinations
Flank region
Minimal target size (based on O. furnacalis PBP Genbank AF133629)
Forward Reverse
1 F1.1 R2.1 exon1,intron 1,exon2 139
2 F1.1 R2.2 exon1,intron 1,exon2 185
3 F1.1 R3.1 exon1,intron 1,exon2,intron2,exon3 381
4 F1.1 R3.2 exon1,intron 1,exon2,intron2,exon3 410
5 F1.2 R2.1 exon1,intron 1,exon2 97
6 F1.2 R2.2 exon1,intron 1,exon2 143
7 F1.2 R3.1 exon1,intron 1,exon2,intron2,exon3 339
8 F1.2 R3.2 exon1,intron 1,exon2,intron2,exon3 368
9 F2.1 R3.1 exon2,intron2,exon3 265
10 F2.1 R3.2 exon2,intron2,exon4 294
11 F2.2 R3.1 exon2,intron2,exon5 215
12 F2.2 R3.2 exon2,intron2,exon6 244
Polyacrilamide gel cutting – Re-PCR
Nested PCR
Amplicon using F2.2-R3.1 primers (Figure 12, lane 11) in standard PCR was used as a template in nested PCR without gel cutting process. This PCR was aimed to reduce non target bands that employed by using internal forward primer F2.1 (see Figure 11) and reverse primer R3.1. Reaction of PCR was performed in 50 µl with other compounds as used in standard PCR included 1 µl amplicon that employed by F2.2-R3.1 primers as a DNA template. Dominant fragments produced by using this PCR method were followed with agarose gel cut, purification and sequencing that were done by sequencing service company.
Touchdown PCR
Amplicon from primer F1.1-R3.1 in standard PCR showed a low concentration single band with 700 bp fragment size (Figure 12, lane 3). This band was strongly expected as PBP gene; however, to make this band can be sequenced, the concentration from this amplicon was needed to be increased. For the purpose to increase DNA concentration for sequence, touchdown PCR was conducted. Touchdown PCR reactions were carried out in thermal cycler following condition: 10 cycles with 97˚C denaturing step for 1 min, 53˚C annealing step for 1 min, and 72˚C extension step for 1,5 min; 10 cycles in which the annealing temperature was reduced to 50˚C; a final 10 cycles using 48˚C annealing temperature. 25 µl volume PCR reactions included 1 µl DNA template, 0,3 mM each dNTP, 0,3 µM each forward and reverse primer, 1x KAPAHIFI fidelity buffer (contain 2 mM Mg2+) and 0,5 Unit KAPAHIFI DNA polymerase (Kapa, South Africa).
Touchdown-Nested PCR
Then 1 µl it amplicon was used in the nested PCR as DNA template. Nested PCR was performed by using internal forward primer from F2.2, F2.1 with reverse primer R3.1, PCR reaction was performed in 25 µl with other compounds as used in Touchdown PCR. However, PCR condition used 50 ˚C temperatures annealing as in nested PCR condition.
DNA Sequencing
Dye labeled ddNTP sequencing method was carried out by sequencing service company. DNA sequence was conducted to two amplicons from two polyacrilamide gel cutting – RePCR method (350 and 300 bp), three from nested PCR amplicons (300,500, and 700 bp), and one from Touchdown-Nested PCR (600 bp) were sequenced.
PCR Product Visualization
Amplicon was separated in 6% polyacrilamide gel electrophoresis (PAGE) using 1X TBE (0.5 M Tris; 0.65 M borac acid; 0.02 M EDTA disodium). It was visualized by sensitive silver staining method (Byun et al. 2009).
Data Analysis
Genetyx software was used for DNA database and translates the DNA sequences into putative amino acid. Homology of S. incertulas PBP with other sequences in Genbank database were explored by using BLASTN and BLASTP (http://blast.ncbi.nlm.nih.gov/ Blast.cgi). Subsequently, sequences were edited using Bioedit and aligned using Clustal X (1.83) software (Thompson et al. 1997). The presence of Open Reading Frame (ORF) was predicted with the HMMgene (v. 1.1) program (http://www.cbs.dtu.dk/services/HMMgene/) (Xiu et al. 2008)
considered to describe the substitution pattern the best. Previous published OR sequences were used as outgroup for tree construction.
[image:50.595.64.508.142.738.2]
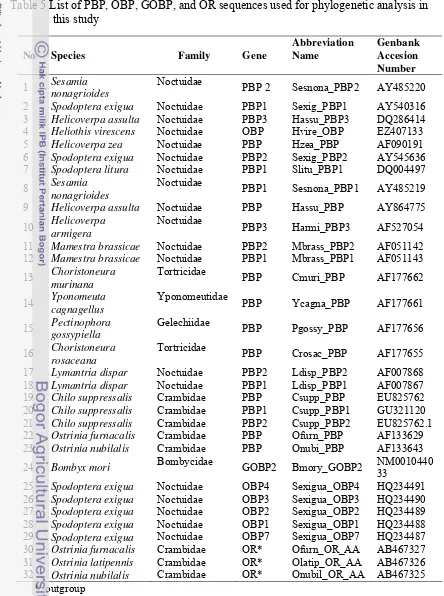
Table 5 List of PBP, OBP, GOBP, and OR sequences used for phylogenetic analysis in this study
No Species Family Gene
Abbreviation Name
Genbank Accesion Number
1 Sesamia
nonagrioides
Noctuidae
PBP 2 Sesnona_PBP2 AY485220
2 Spodoptera exigua Noctuidae PBP1 Sexig_PBP1 AY540316 3 Helicoverpa assulta Noctuidae PBP3 Hassu_PBP3 DQ286414 4 Heliothis virescens Noctuidae OBP Hvire_OBP EZ407133 5 Helicoverpa zea Noctuidae PBP Hzea_PBP AF090191 6 Spodoptera exigua Noctuidae PBP2 Sexig_PBP2 AY545636 7 Spodoptera litura Noctuidae PBP1 Slitu_PBP1 DQ004497
8 Sesamia
nonagrioides
Noctuidae
PBP1 Sesnona_PBP1 AY485219
9 Helicoverpa assulta Noctuidae PBP Hassu_PBP AY864775
10 Helicoverpa
armigera
Noctuidae
PBP3 Harmi_PBP3 AF527054
11 Mamestra brassicae Noctuidae PBP2 Mbrass_PBP2 AF051142 12 Mamestra brassicae Noctuidae PBP1 Mbrass_PBP1 AF051143
13 Choristoneura
murinana
Tortricidae
PBP Cmuri_PBP AF177662
14 Yponomeuta
cagnagellus
Yponomeutidae
PBP Ycagna_PBP AF177661
15 Pectinophora
gossypiella
Gelechiidae
PBP Pgossy_PBP AF177656
16 Choristoneura
rosaceana
Tortricidae
PBP Crosac_PBP AF177655
17 Lymantria dispar Noctuidae PBP2 Ldisp_PBP2 AF007868 18 Lymantria dispar Noctuidae PBP1 Ldisp_PBP1 AF007867 19 Chilo suppressalis Crambidae PBP Csupp_PBP EU825762 20 Chilo suppressalis Crambidae PBP1 Csupp_PBP1 GU321120 21 Chilo suppressalis Crambidae PBP2 Csupp_PBP2 EU825762.1 22 Ostrinia furnacalis Crambidae PBP Ofurn_PBP AF133629 23 Ostrinia nubilalis Crambidae PBP Onubi_PBP AF133643
24 Bombyx mori Bombycidae GOBP2 Bmory_GOBP2 NM0010440 33
25 Spodoptera exigua Noctuidae OBP4 Sexigua_OBP4 HQ234491 26 Spodoptera exigua Noctuidae OBP3 Sexigua_OBP3 HQ234490 27 Spodoptera exigua Noctuidae OBP2 Sexigua_OBP2 HQ234489 28 Spodoptera exigua Noctuidae OBP1 Sexigua_OBP1 HQ234488 29 Spodoptera exigua Noctuidae OBP7 Sexigua_OBP7 HQ234487 30 Ostrinia furnacalis Crambidae OR* Ofurn_OR_AA AB467327 31 Ostrinia latipennis Crambidae OR* Olatip_OR_AA AB467326 32 Ostrinia nubilalis Crambidae OR* Onubil_OR_AA AB467325
4. RESULT
Imagoes Emergence and the Life Cycle Estimation of S. incertulas
Fifty seven S. incertulas imagoes emerged from the first rearing (parental collection from Bogor), consisted of seven and 50 female imagoes. However, in the second rearing using parental collection from Karawang produced more number of imagoes than from the first rearing, i.e. 88 imagoes consisted of 47 males and 41 females. Life cycle span of S. incertulas from the first rearing was longer (48-56 days) than second rearing (44-51 days) (Table 6).
Table 6Life cycle of reared Scirpophagaincertulas
Stage Total inter-stage period
(day)
Inter-stage period (day)
1st rearing 2nd rearing 1st rearing 2nd rearing Imago
0 0 - -
Egg 1 1 1 1
Larva
6 7 6 7
Pupa 41 43 35 36
Visualization of PBP Expected Amplicons of S. incertulas
from Several PCR Modifications
Standard PCR
Among twelve primer combinations were used in standard PCR method (Table 4), primer combinations F1.1-R3.1, F2.1-R3.1, and F2.2-R3.1 showed the expected target bands (see arrow in Figure 12, lane 3, 9 & 11). The size of the expected fragment was shown in primer combination F1.1-R3.1 which was 700 bp (Figure 12, lane 3). Furthermore, there were three expected fragments employed by primer combination F2.1-R3.1 i.e 300, 500, and 700 bp (Figure 12, lane 9). In addition, two expected fragments with size 350 bp and 450 bp were shown using primer combination F2.2-R3.1 (as seen in Figure 12, lane 11).
Polyacrilamide gel cutting – RePCR method
Using primer combinations F2.1-R3.1, one dominant fragment approximately 300 bp was amplified (Figure 13b, lane 9) from multi fragments template (Figure 13a, lane 9) with this method. This method also was able to produce 350 bp dominant fragment (Figure 13b, lane 11) from multi fragments template of standard PCR amplicon by using one pair primer F2.2-R31 (Figure 13a, lane 11)
Nested PCR
[image:53.595.92.491.70.775.2]Primer F2.1 as an internal primer from F2.2 that used in nested PCR gave a same result as in standard PCR by using primer combination F2.1-R3.1 (Figure 12 lane 11). Nested PCR produced more fragments (300, 500, and 700 bp) (Figure 14b) than the amplicon that employed by F2.2-R3.1 as a DNA template (350 bp and 450 bp) (Figure 14a). Gel cutting purification were done for those three amplicons before prior to sequencing process.
Figure 13 Expected PBP amplicons of Scirpophaga incertulas using polyacrilamide gel cutting – RePCR modification. M: 100 bp DNA Ladder, 11: Amplicon F2.2-R3.1, 9: Amplicon F2.1-R3.1, arrow: expected target amplicon
A. Standard PCR (Each sample with two replicates)
B. Polyacrilamide gel cutting – RePCR
(Each sample with three replicates)
Figure 14 Expected PBP amplicons of Scirpophaga incertulas using nested PCR modification. M: 100 bp DNA Ladder, 11: Amplicon F2.2-R3.1from standard PCR that was used as DNA template in Nested PCR, 11’: Amplicon from nested PCR using internal primer F2.1 and reverse primer R3.1, arrow: expected target amplicon
B. Nested PCR using internal primer F2.1
(Each sample with three replicates)
A. Standard PCR
[image:53.595.347.478.543.719.2]Touchdown PCR
Low concentration of 700 bp fragment amplified using F1.1-R3.1 (Figure 15a) can be increased through touchdown PCR (Figure 15b). Nonetheless, concentrations of this fragment still not enough to continue in sequencing process because fragment concentration less than DNA ladder concentration.
Touchdown-nested PCR
Sequences and fragments from nested PCR of amplicon using F2.2-R3.1 primers that was amplified using internal primer F2.1-R3.1 still obtained unsatisfied result (Figure 14b). Therefore, touchdown PCR has been used to produce specific fragments using primer combination F2.2-R3.1 before target fragment was selected by internal primer F2.1 using nested PCR. Three dominant fragments (300, 700, and 1300 bp) were produced by using touchdown PCR with a pair of primer F2.1-R3.1 (Figure 16a). Selection of those fragments that were done by nested PCR gave one promising fragment with high concentration and strongly expected as PBP gene. It has 600 bp fragment size (Figure 16b) that fulfilled minimal target size as shown in Table 3.
Figure 15 Expected PBP amplicon of Scirpophaga incertulas using touchdown PCR modification. M: 100 bp DNA Ladder, 3: Amplicon F1.1-R3.1from standard, 3’: Amplicon F1.1-R3.1 from Touchdown PCR, arrow: expected target amplicon
B. Touchdown PCR
(Each sample with three replicates)
A. Standard PCR
Identification of the PBP Expected Sequences from S. incertulas
There were six sequences obtained from six amplicons; those were produced by using three PCR modification techniques: (i) polyacrilamide gel cutting – rePCR method (two sequences: 217 and 235 bp) (Appendix 1 & 2), (ii) nested PCR (three sequences: 265, 317, one sequence from 700 bp fragment was unreadable) (Appendix 3, 4 & 5), (iii) touchdown-nested PCR (one sequence: 575 bp) (Appendix 6). There were no open reading frame (ORF) found in four sequences from Polyacrilamide gel cutting – RePCR amplicons and nested PCR amplicons. BLASTN result of four sequences from amplicons in point (i) and (ii) did not show the PBP features after aligning those sequences with published PBP sequences in Genbank (Table 7 and Table 8).
The touchdown-nested PCR approach has identified one sequence fragment 575 bp from S. incertulas (named Since_PBPexpected) and contained 169 bp open reading frame (ORF) for a polypeptide of 56 amino acids and an intron of 406 bp (Figure 17). That open reading frame was predicted as exon 3 of PBP expected sequence from S. incertulas and the intron was predicted as intron 2. The 3’end of the intron showed have a typical AG structure (Figure 17).
B. Touchdown-Nested PCR
[image:55.595.134.473.76.285.2](Each sample with three replicates)
Figure 16 Expected PBP amplicons of Scirpophaga incertulas using touchdown- nested PCR modification. M: 100 bp DNA Ladder, 11: Amplicon F2.2-R3.1from touchdown PCR that was used as DNA template in Nested PCR, 11’: Amplicon from Touchdown-Nested PCR using internal primer F2.1 and reverse primer R3.1, arrow: expected target amplicon
A. Touchdown PCR
BLASTP result of 56 amino acids sequence of Since_PBPexpected showed that it has homology structure with GOBP gene of A. mellifera (Table 9) based on alignment of Since_PBPexpected amino acids sequence with amino acids sequence database in Genbank (Figure 18).
Table 7 Top one BLASTN result of two fragments sequenced from Scirpophaga incertulas polyacrilamide gel cutting rePCR amplicon
Sequence size
Accession Description Total
score Max ident E value 241 bp 266 bp AM479556.2 XM_003247 542.1
Vitis vinifera contig VV78X264254.8, whole genome shotgun sequence PREDICTED: Acyrthosiphon pisum
hypothetical protein LOC100569987, transcript variant 1 (LOC100569987), mRNA. 53.6 35.6 92% 100% 5e-04 5.3
Table 8 Top one BLASTN result of two fragments sequenced from Scirpophaga incertulas nested PCR amplicon
Sequence size
Accession Description Total
score Max ident E value 265 bp 317 bp L06178.1 CU633161. 3
Apis mellifera ligustica complete mitochondrial genome
H.numata DNA sequence from clone AEHN-43D2, complete sequence
46.4 42.8 88% 93% 0.00 4 0.05 9
1 TCCCTCACTAAGTCCATAGTGTCGCGCGCTGTGTGTGCCGTAGCGCCGTCTTGTTGAAAC 61
61 CAGATGTGCTCCAGCCCCAATTCGTCCAAACTGGGTATCAAAAACTCATGTAACATGGTT 121
121 CTGTAGCGTTCTCCAGTGACAGTTATCGCATTGCCTCCAGCGTCTTCGAAAAAATAGGGT 181
181 CCGATCACTCTGTCAACACAGATGCCACACCATACAGTAACTTTTAGTGGGTGTAGAGGT 241
241 GTTTCTTGAATAATGCGCGGATTATCAGTGCCCCAGAACCGAGTATTCTGCTTATTGACA 301
301 TGCCCTGAGAGATGGAAATGCGCTTCGTCACTCATAACCAATTTTGAAGGAAAATCATCC 361
361 TCTTCAAGGTTGAGGTTTATGACGCGCTGGCAGTAATCTAGACG
AG
CTCGGTGGTCAGCA 421N L D E L G G Q Q
421 GGCAGCAGCTGATGAGTCATTTGAGTTTTATACGGGAACATCTTTAAGTCTTTCACCAAA 481 A A A D E S F E F Y T G T S L S L S P K
481 ATACGCTGCAGGGTCCGTCTGGAAATGCCCAATTGCGTGGCACGTCGCCTCGTTGATGTT 541 Y A A G S V W K C P I A W H V A S L M F
541 TCTGGACACTCTTCCACATCTTCGCGCACAGCAT 575 L D T L P H L R A Q H
Phylogenetic Analysis of Third Exon PBP Sequence of S. incertulas
I aligned the putative amino acid sequences of third exon region from Since_PBPexpected with published amino acid sequences of third region PBPs, from various species as in Table 5. Lepidoteran PBPs have three conserved residues motifs i.e Alanine (Ala8), Cysteine (Cys34), Leucine (Leu43) and Since_PBPexpected possessed one conserved cysteine that might form disulfide bridge, and the hydrophobic domains (Figure 19).
Alignment of third exon putative amino acid sequencesPBPs among species showed there were three conserved cysteine residues, however, Since_PBPexpected amino acid only showed one concerved cysteine (Figure 20). Since_PBPexpected can be classified into PBP group by phylogenetic analysis (Figure 21). Since_PBP expected putative amino acid sequences has the longest distance with Sexigua_OBP1 (distance = 6,536) and Sexigua_OBP2 (distance = 6,437); Moreover, it has nearest distance with Ofurn_PBP (distance = 3.366) and Onubu_PBP (distance = 3,366) (Table 10).
Table 9 BLASTP result from third exon amino acid sequences of Scirpophaga incertulas touchdown-nested PCR amplicon
Accession Description Identities E value
1TUJ_A Chain A, Solution Structure Of The Honey Bee General Odorant Binding Protein Asp2
35% 0,032
pdb|1TUJ|A Chain A, Solution Structure Of The Honey Bee (Apis mellifera) General Odorant Binding Protein Asp2 In Complex With Trimethylsilyl-D4 Propionate Length=123
Score = 20.0 bits (40), Expect = 0.032, Method: Compositional matrix adjust. Identities = 9/26 (35%), Positives = 12/26 (46%), Gaps = 0/26 (0%)
Query 1 LGGQQAAADESFEFYTGTSLSLSPKY 26 LG +A + E GT L + P Y Sbjct 47 LGCLKACVMKRIEMLKGTELYVEPVY 72
Figure 20 Conserved cysteine amino acid position in exons of three moths PBP gene and the schematic of intron and exon size from three moths PBP gene. C: Cysteine amino acid; O. furnacalis (Willet and Harrison 1999), S. exigua (Xiu and Dong 2007), S. incertulas (present study). Figure 19 Alignment of the amino acid sequences from translated third exon of
Scirpophaga incertulas PBP expected. Conserved amino acids are shaded yellow, “ ”: conserved cysteine, PBP names are the same as inTable 5.
Table 10 Genetic distance of Scirpophaga incertulas PBP amino acid sequence with published GOBP, OBP, and OR amino acid sequence from various species in Lepidopteraa
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18
1 Sesnona_PBP2
2 Sexig_PBP1 0.827
3 Hassu_PBP3 1.128 0.432
4 Hvire_OBP 2.952 2.179 2.171
5 Hzea_PBP 0.398 0.685 1.002 2.284
6 Sexig_PBP2 0.561 0.723 0.962 2.550 0.166
7 Slitu_PBP1 0.749 0.087 0.392 2.147 0.594 0.632
8 Sesnona_PBP1 0.699 0.473 0.598 2.203 0.665 0.709 0.429
9 Hassu_PBP 0.421 0.761 1.029 2.317 0.094 0.239 0.667 0.695
10 Harmi_PBP3 1.128 0.432 0.000 2.171 1.002 0.962 0.392 0.598 1.029
11 Mbrass_PBP2 0.412 0.749 1.144 2.874 0.186 0.274 0.656 0.797 0.221 1.144
12 Mbrass_PBP1 0.722 0.548 0.608 1.963 0.741 0.812 0.504 0.159 0.783 0.608 0.875
13 Cmuri_PBP 0.897 0.929 0.861 3.140 0.539 0.477 0.860 0.736 0.554 0.861 0.671 0.830
14 Ycagna_PBP 0.697 0.351 0.473 1.893 0.638 0.731 0.317 0.506 0.722 0.473 0.817 0.456 0.730
15 Pgossy_PBP 0.845 0.744 0.935 2.545 0.416 0.585 0.740 0.835 0.435 0.935 0.510 0.772 0.414 0.682
16 Crosac_PBP 0.881 0.852 0.926 2.851 0.525 0.578 0.787 0.742 0.549 0.926 0.657 0.774 0.202 0.724 0.365
17 Ldisp_PBP2 0.669 1.228 1.080 2.354 0.649 0.724 1.167 0.785 0.657 1.080 0.584 0.780 0.919 1.070 0.840 0.815
18 Ldisp_PBP1 0.673 1.105 1.308 2.286 0.700 0.764 1.095 0.996 0.712 1.308 0.742 1.067 0.709 1.003 0.732 0.686 0.676
19 Csupp_PBP 1.077 0.820 1.045 2.525 0.874 0.881 0.977 1.017 0.865 1.045 1.055 0.890 0.683 0.917 0.518 0.521 0.977 0.730
20 Csupp_PBP1 0.893 0.752 1.043 2.706 0.877 0.901 0.792 0.951 0.985 1.043 0.850 0.807 0.926 0.970 0.622 0.752 0.939 0.755
21 Csupp_PBP2 1.077 0.820 1.045 2.525 0.874 0.881 0.977 1.017 0.865 1.045 1.055 0.890 0.683 0.917 0.518 0.521 0.977 0.730
22 Ofurn_PBP 0.658 0.719 1.034 2.407 0.430 0.544 0.716 0.912 0.415 1.034 0.602 0.847 0.534 0.777 0.391 0.525 0.936 0.652
23 Onubi_PBP 0.658 0.719 1.034 2.407 0.430 0.544 0.716 0.912 0.415 1.034 0.602 0.847 0.534 0.777 0.391 0.525 0.936 0.652
24 Bmory_GOBP2 1.610 1.172 1.178 2.618 1.599 1.592 1.059 1.386 1.471 1.178 1.657 1.460 1.257 1.219 1.382 1.194 1.599 1.611
25 Sexigua_OBP4 3.407 3.058 2.826 1.324 3.098 2.970 3.202 3.313 3.252 2.826 3.358 3.046 3.685 2.319 3.023 3.668 3.150 3.322
26 Sexigua_OBP3 2.708 2.443 2.212 1.106 2.693 2.689 2.265 2.069 2.853 2.212 3.001 1.972 3.298 2.148 2.560 3.215 2.113 2.823
27 Sexigua_OBP2 3.376 3.038 2.809 2.247 3.122 3.015 3.162 2.652 2.967 2.809 3.584 2.730 3.277 3.154 2.816 3.265 3.368 3.551
28 Sexigua_OBP1 2.894 2.648 2.612 0.561 2.447 2.655 2.705 2.688 2.656 2.612 2.950 2.406 3.626 2.353 2.958 3.340 2.408 2.624
29 Sexigua_OBP7 2.642 2.838 2.754 2.758 2.283 2.005 2.880 2.043 2.351 2.754 2.607 2.421 2.290 2.435 2.645 2.316 2.118 2.466
30 Ofurn_OR_AA 3.049 3.489 3.556 2.429 3.073 2.926 3.539 3.322 3.055 3.556 2.855 3.044 2.713 3.626 3.043 2.490 3.326 2.911
31 Olatip_OR_AA 2.964 3.569 3.426 2.426 3.113 2.966 3.617 3.337 2.880 3.426 2.900 3.166 2.627 3.492 2.889 2.412 3.295 2.679
32 Onubil_OR_AA 3.049 3.489 3.556 2.429 3.073 2.926 3.539 3.322 3.055 3.556 2.855 3.044 2.713 3.626 3.043 2.490 3.326 2.911
33 Since_PBPexpect 4.725 4.338 5.108 4.747 4.249 4.345 4.493 4.753 4.002 5.108 4.547 4.532 4.815 4.649 4.766 4.645 4.356 4.736
19 20 21 22 23 24 25 26 27 28 29 30 31 32
20 Csupp_PBP1 0.801
21 Csupp_PBP2 0.000 0.801
22 Ofurn_PBP 0.581 0.637 0.581
23 Onubi_PBP 0.581 0.637 0.581 0.000
24 Bmory_GOBP2 1.412 1.167 1.412 1.221 1.221
25 Sexigua_OBP4 3.103 3.838 3.103 3.261 3.261 2.688
26 Sexigua_OBP3 2.655 2.232 2.655 2.783 2.783 2.033 1.828
27 Sexigua_OBP2 2.584 3.101 2.584 2.807 2.807 2.938 2.166 2.423
28 Sexigua_OBP1 2.963 2.738 2.963 2.657 2.657 2.551 1.867 0.737 3.096
29 Sexigua_OBP7 2.537 3.362 2.537 2.498 2.498 2.811 2.102 2.822 3.756 2.838
30 Ofurn_OR_AA 2.737 2.472 2.737 3.117 3.117 3.637 3.466 4.030 3.883 3.399 3.401
31 Olatip_OR_AA 2.584 2.452 2.584 2.967 2.967 3.449 3.293 4.033 3.871 3.391 3.434 0.058
32 Onubil_OR_AA 2.737 2.472 2.737 3.117 3.117 3.637 3.466 4.030 3.883 3.399 3.401 0.000 0.058
33 Since_PBPexpect 3.880 5.541 3.880 3.366 3.366 5.934 3.660 6.437 3.973 6.536 5.010 5.318 5.365 5.318
a
Name of species refer to Table 5. Underlined bold font indicates the species that have nearest amino acid sequence genetic distance with S. incertulas,