The influence of glutathion S-transferase P-1 polymorphism A313G
rs1695 on the susceptibility to cyclophosphamide hematologic
toxicity in Indonesian patients
Keywords: breast cancer, cyclophosphamide, GSTP1 polymorphism, hematology toxicity
pISSN: 0853-1773 • eISSN: 2252-8083 • http://dx.doi.org/10.13181/mji.v25i2.1308 • Med J Indones. 2016;25:118–26 • Received 09 Nov 2015 • Accepted 15 May 2016
Corresponding author: Dita Hasni, [email protected]
Copyright @ 2016 Authors. This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial 4.0 International License (http://creativecommons.org/licenses/by-nc/4.0/), which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original author and source are properly cited.
Dita Hasni,1,2 Kamal B. Siregar,3 Hadyanto Lim1,4 1 Biomedical Master Program, Faculty of Medicine, Universitas Sumatera Utara, Medan, Indonesia
2 Departement of Pharmacotherapy, Faculty of Medicine, Universitas Baiturrahmah, Padang, Indonesia
3 Departement of Surgery, Faculty of Medicine, Universitas Sumatera Utara, Medan, Indonesia
4 Departement of Pharmacology, Faculty of Medicine, Universitas Methodist Indonesia, Medan, Indonesia Clinical Research
ABSTRAK
Latar belakang: Kemoterapi sering menimbulkan efek samping berupa toksisitas hematologi. Derajat toksisitas yang timbul
sering dikaitkan dengan polimorfisme genetik. Penelitian ini bertujuan mengetahui apakah polimorfisme genetik GSTP1 A313G yang berperan pada detoksifikasi siklofosfamid dapat
memprediksi insidensi dan derajat toksisitas hematologi siklofosfamid.
Metode: 91 perempuan Indonesia yang telah didiagnosis kanker payudara di RSUP Haji Adam Malik, Medan dan diterapi dengan regimen siklofosfamid, doxorubicin/epirubicin, dan 5-FU diikutsertakan pada penelitian kohort retrospektif ini.
DNA diekstraksi dari leukosit perifer pasien dan dilanjutkan
dengan pemeriksaan genotipe GSTP1 A313G dengan metode
polymerase chain reaction-restriction length fragment polymorphism (PCR-RFLP). Distribusi frekuensi genotipe dan alel ditentukan dengan Hardy-Weinberg Equilibrium. Data derajat toksisitas hematologi (leukopenia dan neutropenia
pasca-kemoterapi siklus 1 dan 3) dikumpulkan dari rekam
medis pasien dan dianalisis dengan uji Kai kuadrat. Antar kelompok genotipe yang berbeda.
Hasil: Didapatkan proporsi GSTP1 A313A (wild type) 60,4%, GSTP1 A313G (heterozigot mutan) 29,7%, dan GSTP1 G313G (homozigot mutan) 9,9%. Tidak ada penyimpangan frekuensi genotipe dan alel yang signifikan terhadap Hardy-Weinberg Equilibrium. Ditemukan pasien dengan alel G (A/G&G/G)
cenderung mengalami leukopenia dengan derajat yang lebih berat dibandingkan dengan pasien GSTP1 wild type alel (A/A)
pasca-kemoterapi siklus ke-3 (p<0,05).
Kesimpulan: Polimorfisme GSTP1 exon 5 A313G memiliki
hubungan dengan derajat toksisitas hematologi pada pasien kanker payudara yang mendapat kemoterapi siklofosfamid.
ABSTRACT
Background: Chemotherapy often causes side effects such as hematologic toxicity. The degree of toxicity is often associated with genetic polymorphism. This study aims to determine the influence of GSTP1 A313G polymorphism, an enzyme responsible for detoxifying cyclophosphamid, on incidence and severity of cyclophosphamid hematologic toxicity.
Methods: 91 Indonesian females diagnosed with breast cancer at Haji Adam Malik Central General Hospital, Medan, receiving cyclophosphamide, doxorubicin/epirubicin and 5-FU were included in this retrospective cohort study. DNA was extracted from peripheral leukocytes and GSTP1 A313G genotyping was analyzed using polymerase chain reaction-restriction length fragment polymorphism (PCR-RFLP). Genotype deviation and allele frequencies were also determined by Hardy-Weinberg Equilibrium. The degrees of hematologic toxicity (leucopenia and neutropenia data after chemotherapy cycles 1 and 3) were collected from the patient medical records. The data were analyzed using chi-square test.
Results: 60.4% of the patients had the wildtype (A/A), while 29.7% were heterozygous (A/G), and 9.9% were homozygous mutant (G/G). There was no significant deviation of allele and genotype frequency from Hardy-Weinberg Equilibrium. The G allele (A/G & G/G) contributes to more severe degree of leukopenia compared to patients with wild type allele (A/A) (p<0.05) after the 3rd chemotherapy cycles.
Cytotoxic chemotherapy is one of the modalities in breast cancer treatment, whether it is a neoadjuvant or adjuvant that aims to downstage, downsize and prevent metastasis to other organs.1 Cytotoxic chemotherapy, on the other
hand, has side effects such as neutropenia and leucopenia, since the chemotherapeutic agents work indifferently on cancer cells and on normal cells which actively proliferate, such as blood cells.2
Cyclophosphamide is one of the leading drugs for combination cytotoxic chemotherapy regimen in breast cancer treatment. Cyclophosphamide is a prodrug that requires phase I biotransformation for the activation of the cytotoxic active metabolite being mediated by CYP450 enzyme.
Detoxification of cyclophosphamide requires
phase II biotransformation mediated by glutathione S-tranferase 1 (GSTP1) enzyme by catalyzing the active metabolite with glutathione to form a non-toxic water soluble metabolite.3
GSTP1 is an important enzyme in detoxifying cyclophosphamide metabolites. This enzyme is expressed by GSTP1 gene which is located on the
chromosome 11q13 and has non-synonymous
nucleotide polymorphism at exon 5 at nucleotide
313 (SNP’s ID:rs1695) where adenine changes
to guanine.4 This causes the substitution of
amino acid from isoleucine to valine at codon
105 which results in lower affinity between GSTP1 and its substrate up to 30% and affects
the catalytic activity of GSTP1 during phase II biotransformation where the active metabolite is catalyzed to a non-toxic water soluble form.5
Zhang et al6 found that GSTP1 variant 105Val has
lower temperature stability which is influenced
by catalytic activity of its substrates. Patients with homozygous isoleucine (Ile/Ile) have the highest GSTP1 enzyme activity and patients with heterozygous Ile/Val have a lower enzymatic activity compared to the homozygous mutant Val/Val.6 A study done by Zhong et al5 also found
that GSTP1 activity in the erythrocyte was two to three times lower in GSTP1 Val/Val compared to Ile/Ile in various age groups.5
The polymorphism effect on GSTP1 enzymatic activity will have a downstream effect on activity
of the cyclophosphamide detoxification process
and its toxic metabolites (phosphoramide mustard and acrolein). This will accumulate
toxic metabolites in the body which increases the risk of hematologic toxicity then severity in chemotherapy containing cyclophosphamide afterward.6,7 Tran et al8 reported that CYPB26,
GSTA1, GSTP1 polymorphisms were related to incidence of severe hematologic toxicity in glomerulonephritis patients who receive cyclophosphamide therapy.8,9 Similar
observations were also seen in studies done by Zhang et al6 and Artini et al10 which showed
thatbreast cancer patients with GSTP1 Ile/Val and Val/Val polymorphisms has a tendency to experience the heavier degree of hematologic toxicity after chemotherapy compared to the wild type (Ile/Ile).6,10
To sum up the literature review, it is an urgency to downsize the number of drug-induced hematological toxicity especially leucopenia and neutropenia in our population from who were diagnosed breast cancer receiving cyclophosphamide, adriamycin/epirubicin and
fluorouracyl regimens (CAF/CEF). This study conducted by aiming to determine the influence
of genetic variance in the GSTP1 gene involved
in detoxification of cyclophosphamide active
metabolite could affect susceptibility on the incidence and severity of hematologic toxicity after cyclophosphamide chemotherapy.
METHODS
Patient selection
From July 2013 to February 2014, 91 Indonesian
women were recruited in this retrospective cohort study. Subjects were selected from surgical oncology outpatient clinic of Adam Malik Hospital,
Medan, Indonesia who fulfilled elligibility criteria
in terms of a histopathological diagnosis as gold standard of breast cancer diagnosis which had been hospitalized and treated with a combination of cyclophosphamide, doxorubicin/epirubicin
and 5-fluorouracyl (CAF/CEF). These regimens
comprise 500 mg/m2 of cyclophosphamide, 50
mg/m2 adriamicyn or 50 mg/m2 epirubicin, and
500 mg/m2 5-FU intravenously on day one of
and immunosuppressant less than two months prior to chemotherapy. Then, patients were recruited by consecutive sampling with inclusion and exclusion criteria set above. The protocol of this study was approved by Medical Faculty of Universitas Sumatera Utara Ethical Committee
(No. 322/KOMET/FK USU/2013).
Demographic, clinico-pathological data and blood samples were collected from patient notes and medical record at medical oncology outpatient clinic where patients attended to treatment or follow-up regularly.
Genomic DNA isolation
After written informed consent was obtained from all participants, blood sample from each patient was collected into ethylenediaminetetraacetic acid
(EDTA) containing storage tube and stored at -20°C before deoxyribonucleic acid (DNA) extraction,
then genomic DNA was purified from peripheral
blood leucocyte using the Wizard® genomic DNA
purification kit (Promega, USA) with procedure commonly used in integrated laboratory of Medical Faculty of Universitas Sumatera Utara
An amount of 1 mL of peripheral blood containing EDTA was transferred to Eppendorf tube and centrifuged in Eppendorf centrifuge
5,430 at speed of 3,000 RPM for 15 minutes at
room temperature. The pellet-leucocyte form
was pipetted as many as 300µL transferred to Eppendorf tube containing 900µL of red blood
cell (RBC) lysis solution, then inverted three
times and incubated at 3-5°C for 10 minutes. The
sample was then centrifuged in centrifuge 5,702R
at a speed of 13,000 RPM for three minutes at
room temperature. The supernatant was removed by leaving the pellet in the form of mononuclear leucocytes.
The purification were repeated three times to
obtain a white pellet, named as consideration the absence of red blood cells. Then the white
pellet was added 300µL of nuclei lysis solution
(NLS) and inverted until it became a homogenous
solution. Next the solution added 100µL of protein
precipitation solution and vortexed with an auto vortex (Biosan V-1 Plus) for 20 seconds. The lysate
was centrifuged at 13,000 RPM for one minute at
room temperature. Then, the supernatant was
poured into a new Eppendorf contained 300µL
isopropanolol. The tube was inverted 25 times
until it formed a collection of DNA strands. Then
the samples were centrifuged at 13,000 RPM for
one minute at room temperature, so that DNA-containing precipitate appeared white.
The supernatant was removed and then added 1 ml of cold 70% ethanol, this mixture was then
centrifuged at 13,000 RPM for one minute at room
temperature. The supernatant was discarded and the DNA dried in open air by reversing the tube
for one hour. DNA was rehydrated with 100µL
DNA rehydration solution and then incubated at 4°C overnight. DNA yields were not estimated their concentration. Then they were stored at -80°C until all of samples collected.
Genotyping of GSTP1 single nucleotide polymorphism
The amplification of isolated DNA was performed
with polymerase chain reaction (PCR) with each reaction containing DNA GoTaq® Green Master
Mix (Promega), the sequences of the forward and
reverse primers for GSTP1 exon 5 were 5’-GAG GAA ACT GAG ACC CAC TGAG-3’ and 5’-AGC CCC TTT CTT TGT TCA GCC-3’,10,11 nuclease free water,
and DNA template with the final volume of 25μL.
The DNA chain was denaturated by incubation
at 94°C for five minutes, followed by 35 series of chain reaction (94°C for 30s, then annealed at 61°C for 30s, and 72°C for 30s) followed by final expansion step at 72°C for five minutes.
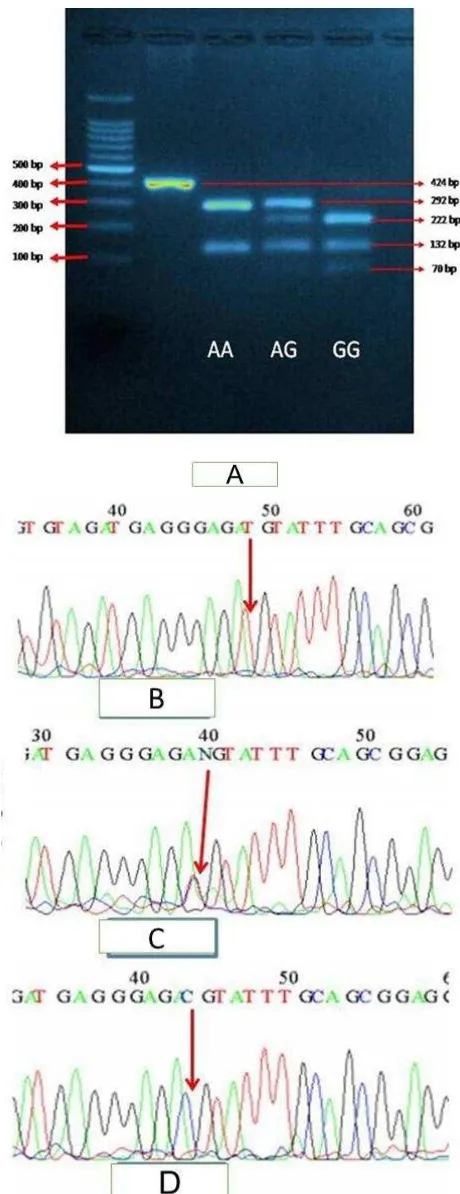
The PCR products were then visualized using 2% agarose gel electrophoresis stained with ethidium
bromide and a DNA fragment at 424 bp confirmed the amplification. The PCR products were then
subjected to enzyme restriction with BsmA1 and
incubated for 30 minutes at 55°C followed by visualization with 3% agarose gel electrophoresis
stained with ethidium bromide. The wild type
(A/A) allele showed fragments at 292 and 132 bp,
the heterozygous allele (A/G) showed fragments
at 292, 222, 132, and 70 bp and the mutant (G/G) showed fragments at 222, 132, and 70 bp.
DNA sequencing
To validate data generated by polymerase chain reaction-restriction length fragment polymorphism (PCR-RFLP) assay, 10% of the
samples of unpurified PCR product were randomly
Assessment of hematological toxicities
The degree of hematological toxicity was evaluated from leukocyte and neutrophil count on day 19 after chemotherapy cycles one and three based on common terminology criteria for
adverse events (CTCAE v.3.0).12
Degree of leucopenia (grade I=<4,000-3,000/
mm3, grade II=<3,000-2,000/mm3, grade III=<2,000-1,000/mm3, and grade IV=<1,000/ mm3). Degree of neutropenia (grade I=<2,500-1,500/mm3, grade II=<1,500-1,000/mm3, grade III=<1,000-500/mm3, grade IV=<500/mm3).
Statistical analysis
Qualitative variables were summarized by their frequency distribution and quantitative variables by their mean and standard deviation (SD) or median and interquartile range.
GSTP1 genotypes were tested as to whether they were distributed according to Hardy-Weinberg Equilibrium. Chi square test for deviation from Hardy Weinberg equilibrium was used to estimate
differences in allele frequencies. The influence
between GSTP1 genotype with cyclophosphamide-induced leucopenia and neutropenia after chemotherapy series one and three were estimated using p value. A p<0.05 was considered statistically
significant.
RESULTS
Demography characteristics and subjects clinicopathology
Total of 91 patients with the avarage age of
52.53±8.79 years (range 30–76 years) were evaluated in this study. The subjects consist of several ethnic groups consisting of 41 Bataknese
patients (45.1%), 30 Javanese patients (33.3%),
nine Acehnese patients (9.9%), eight Malay (8.8%),
and the three rest patients (3.35%) consists of
Chinese, Dayaknese, and Indian ethnics. The clinico-pathological features of the subjects and variant genotype could be seen in Table 1.
DNA amplification to detect GSTP1 polymorphism
Interpretation of RFLP product for the A313G on
424 bp PCR fragment with BsmA1 were shown by different pattern on gel electrophoresis (Figure 1). Digestion of the 424 bp fragment of
Subject characteristics All subjects n (%)
GSTP1 A313G genotype
AA n (%)
AG n (%)
GG n (%)
Total 91 55 27 9
Ethnicity
Javanese 30 (33.0) 19 (63.3) 10 (33.3) 1 (3.3)
Bataknese 41 (45.1) 20 (48.8) 14 (34.1) 7 (17.1)
Malay 8 (8.8) 7 (87.5) 0 1 (12.5)
Acehnese 9 (9.9) 6 (66.7) 3 (33.3) 0
The others 3 (3.3) 3 (100) 0 0
Age (years, mean±SD) 52.53±8.79 53.16±8.57 50.74±8.41 54.00±11.28 Breast cancer stage
IIa 9 (9.9) 8 (88.9) 1 (11.1) 0
IIb 23 (25.3) 14 (60.9) 7 (30.4) 2 (8.7)
IIIa 7 (7.7) 1 (14.3) 4 (57.1) 2 (28.6)
IIIb 47 (51.6) 28 (59.6) 14 (29.8) 5 (10.6)
IIIc 4 (4.4) 3 (75.0) 1 (25.0) 0
IV 1 (1,1) 1 (100.0) 0 0
Histopathology
Invasive ductal carcinoma 77 (84.6) 49 (63.6) 20 (26.6) 8 (10.4) Invasive lobular carcinoma 12 (13.2) 5 (41.7) 7 (58.3) 0
The others 2 (2.2) 1 (50.0) 0 1(50.0)
Histopathological degree
Grade I 22 (24.2) 12 (54.5) 10 (45.5) 0
Grade II 34 (37.4) 22 (64.7) 6 (17.6) 6 (17.6)
Grade III 21 (23.1) 12 (57.1) 8 (38.1) 1 (4.8)
Unknown 14 (15.4) 9 (64.3) 3 (21.4) 2 (14.3)
Chemotherapy
Neoadjuvant 57 (62.6) 34 (59.6) 15 (26.3) 8 (14.0)
Adjuvant 34 (37.4) 21 (61.8) 12 (35.3) 1 (2.9)
Chemotherapy regimen
CAF 79 (86.8) 47 (59.5) 24 (30.4) 8 (10.1)
CEF 12 (13.2) 8 (66.7) 3 (25.0) 1 (8.3)
Table 1. Demographic and clinicopathological characteristics in various genotype subjects
GSTP1= glutathion S-Transferase P-1; CAF= cyclophosphamide adriamycin fluorouracyl; CEF= cyclophosphamide epirubicin fluorouracyl
313 AA genotype gave two fragments of 292 and 132 bp, the 313 AG genotype resulted in four fragments at 292, 222, 132, and 70 bp, whereas the 313 GG genotype result in three fragments at 222, 132, and 70 bp. The PCR-RFLP results were confirmed with DNA sequencing.
Distribution of GSTP1 polymorphism
The genotype distribution of the patients is shown on Figure 2 which demonstrates that 55 patients (60.4%) had the wild type (A/A), while 27 patients (29.7%) were heterozygous (A/G), and nine patients
(9.9%) were homozygous mutant (G/G). The allele frequencies of single nucleotide polymorphism
(SNPs) A313G GSTP1 gene on this groups were 45 (24.73%) of G allele, which is considered as minor allele frequency and 137 (75.27%) of A allele classified as the major allele frequency.
In this study, we compared the distribution of GSTP1 genotype and allele frequency in various
populations including Indonesian, South Korean,
Figure 2. Distribution of genotype GSTP1 A313G polymor -phism on subjects. The genotype distributions of the patients were as follows, 60.4% of the patient has the GSTP1 313AA (wild type), while 29.7% is GSTP1 313AG (heterozygous), and 9.9% is GSTP1 313GG (homozygous mutant)
results are shown in Table 2. The distribution
of GSTP1 A313G genotypes in our data showed that it was not significantly deviated from
Hardy-Weinberg equilibrium (p>0.05).
Hematologic toxicity incidence
The incidence of hematologic toxicity was evaluated from neutropenia and leucopenia after chemotherapy. The incidence of neutropenia in this study after cycle one chemotherapy was
Population No of individuals
GSTP1 Genotype [n, (%)] Allele Frequency [n, (%)] p
AA AG GG A G
Medan, Indonesia (present study) 91 55 (60.4) 27 (29.7) 9 (9.9) 137 (75.27) 45 (24.73) 0.0529 Javanese 30 19 (63.3) 10 (33.3) 1 (3.3) 48 (80) 12 (20) 0.8195 Bataknese 41 20 (48.8) 14 (34.1) 7 (17.1) 54 (65.85) 28 (34.15) 0.1232
Malay 8 7 (87.5) 0 1 (12.5) 14 (87.5) 2 (12.5) 0.0047*
Acehnese 9 6 (66.7) 3 (33.3) 0 15 (83.33) 3 (16.67) 0.5485
The others 3 3 (100) 0 0 6 (100) 0 0.6442
Yogyakarta, Indonesia10 50 25 (50) 20 ( 40) 5 (10) 70 (70) 30 (30) 0.7363
Kazakhstan16 120 63 (52.1) 46 (38.7) 11 (9.2) 172 (71.67) 68 (28.33) 0.5389
North China7 920 540 (58.7) 325 (35.3) 55 (6.0) 1405 (76.36) 435 (23.64) 0.5132 Taiwan17 192 122 (64.2) 65 (33.9) 5 (1.9) 309 (80.89) 73 (19.11) 0.1637 Brazil 750 330 (44) 329 (44) 91 (12) 989 (65.93) 511 (34.07) 0.5198 Caucasian18 599 268 (45) 270 (45) 61 (10) 806 (67.28) 392 (32.72) 0.5608 African American 151 62 (41) 59 (39) 30 (20) 183 (60.6) 119 (39.4) 0.0255* Italy19 48 22 (45.8) 22 (45.8) 4 (8.3) 66 (68.75) 30 (31.25) 0.6442 Table 2. Distribution of GSTP1 genotypes and allele frequency by Hardy-Weiberg Equilibrium on breast cancer patients in vari-ous population
* Significant; GSTP1= glutathion S-transferase P-1
Degree of hematological toxicity
After first cycle n (%)
After third cycle n (%)
Neutropenia 35 (38.5) 54 (59.4) Grade I 22 (24.2 ) 28 (30.8) Grade II 9 (9.9 ) 13 (14.3) Grade III 2 (2.2) 6 (6.6)
Grade IV 2 (2.2) 7 (7.7)
Leucopenia 11 (12,1) 33 (36.3) Grade I 8 (8.8 ) 11 (12.1)
Grade II 3 (3.3) 18 (19.8)
Grade III - 3 (3.3)
Grade IV - 1 (1.1)
Table 3. Degree of hematological toxicity after first and third cycles chemotherapy
38.5% and increased to 59.4% after chemotherapy
cycle three. The incidence of leucopenia after the
first cycle chemotherapy in this study was 12.1% and increased to 36.3% after the third cycle chemotherapy (Table 3).
GSTP1 polymorphism and hematologic toxicity
This study did not found difference in degree of neutropenia after CAF/CEF chemotherapy cycle
one between the GSTP1 313AA and the GSTP1
This study reported did not found difference in degree of leucopenia after CAF/CEF chemotherapy
cycle one between the GSTP1 313AA and the GSTP1 313AG+GG genotypes but a significant
difference in degree of leucopenia was found after cycles three chemotherapy (p<0.05) (Table 4 and Table 5).
DISCUSSION
Cyclophosphamide is a cytotoxic agent used for breast cancer treatment which can suppress
Hematological toxicity,
Neutropenia 31 4 0.546*
AA 22 (70.96) 4 (100)
AG 8(25.81) 0 (0)
GG 1(3.23) 0 (0)
AG and GG 9 (29.04) 0 (0) 0.286†
Leucopenia 11 0 N/A
AA 9 (81.82) 0 thion S-transferase P-1; N/A= statistical test not applicable; AA= adenine adenine; GG= guanine guanine; AG= adenine guanine
Neutropenia 41 13 0.772*
AA 29 (70.73) 6 (46.15)
AG 8 (19.51) 6 (46.15)
GG 4 (9.76) 1(7.69)
AG and GG 12 (29.27) 7 (53.85) 0.106†
Leucopenia 29 4 0.035a
AA 22(75.86) 0
AG 6 (20.69) 3 (75.0)
GG 1 (3.45) 1 (25.0)
AG and GG 7(63.6) 4 (100) 0.004‡
Table 5. Degree of hematological toxicity after third cycle chemotherapy in GSTP1 genotype
* Kolmogorov-Smirnov test; † pearson chi square test; ‡ fisher
exact test; GSTP1= glutathion S-transferase P-1
cancer and normal cell such as blood cells since these cells are actively proliferating.2 Blood cells
with the shortest life span will firstly experience
suppression and followed by other cells with the order of neutrophils, leucocytes, platelets and erythrocytes.2,13
In this study, 38.5% and 59.4% of breast cancer patients suffered neutropenia after first and third
cycles of CAF/CEF chemotherapy respectively. Previous study by Han et al14also showed similar
results. In early stage breast cancer, 52.5% patients who received CEF adjuvant chemotherapy experienced neutropenia with mild neutropenia (degree one and two) of 41.5% and severe degree (degree three and four) of 11%.14
The incidence of neutropenia after first cycle
chemotherapy in this study is higher than
previous study done by Keswara et al13 which
shows the incidence of neutropenia 27.4% after 12 days of cycle one CAF chemotherapy.13 his difference might be due to difference in time of observation. In this study, neutropenia occurred
on day 19 whilst in the study of Keswara et al13 it
took place on days seven and 12.
In this study, 11.1% and 36.3% of breast cancer patients suffered from leucopenia after first and
third cycles CAF/CEF chemotherapy, respectively. The results after third cycle chemotherapy closely resemble previous study by Artini et al10
which showed 38% of breast cancer patients receiving cyclophosphamide in taxan-based and antracyclin-based regimens, experienced leucopenia after treated overall cycles of chemotherapy.10 The incidence of leucopenia in
this study, however, was much lower than the study done by Palappallil et al15 in breast cancer
patients receiving CAF regimen. These authors reported that 66% and majority of the subject only showed degree one leucopenia.15 The difference
in incidence of leucopenia between this study and study of Palappallil et al15 may be attributed
to difference in observation times: cycles one and three in this study vs all cycles in this study.
In this study, genotype counts did not significantly
deviate from those expected from the
Hardy-Weinberg equilibrium (p=0.053). This finding
is similar to the Yogyakarta, Indonesian,10
Khazakstan,16 Chinese,7 Taiwanese,17 Brazilian,18
data showed a significant deviated from
Hardy-Weinberg equilibrium. This result is similar to African American race from Brazilian population.18
The present study investigates the influence
of GSTP1 polymorphism on susceptibility of hematological toxicity induced by
cyclophosphamide. Our results showed there was
no difference in degree of leucopenia between GSTP1 genotypes after chemotherapy cycle one, which is in contrast with that after cycle three of
chemotherapy. GSTP1 313AG+313GG genotype
had higher incidence of three to four degree
leucopenia than GSTP1 313AA genotype (p=0.035
and p=0.004). This may be due to the accumulation of cycloposphamide and its metabolites due to decreased activity of detoxifying enzymes GSTP1
for the GSTP1 313AG+GG genotype.
Our results also found that after cycles one and three chemotherapy, there were no differences in the degree of neutropenia between the GSTP1
313AA and The GSTP1 313AG+GG genotypes.
The situation is different with the incidence of leukopenia, probably due to the shorter life cycle of neutrophil than that of leukocytes. So at day 19 post chemotherapy blood neutrophil levels have returned to normal
Therefore, it is concluded that there is an association between GSTP1 polymorphism and hematological toxicity induced by
cyclophosphamide. This finding is similar to
previous study which showed higher incidence of
severe hematologic toxicity in GSTP1 313AG+GG patients compared to GSTP1 313AA. This is in contrast to the findings of Ekhart et al9 which
stated that GSTP1 polymorphism had no influence
on inter-individual cyclophosphamide and 4-hydroxycyclophosphamide pharmacokinetics.9
Ekhart et al20 stated that GSTP1 polymorphism
cannot be the contributing factor to variations of toxicological response.
In conclusion, our data indicated that host constitutional variability such as GSTP1 polymorphism played a role in the severity of hematologic toxicity in patients receiving cyclophosphamide in CAF/CEF regimen. It will be interesting to determine the polymorphism of metabolic enzymes involved in phase I and phase II biotransformation, especially drugs involved in the chemotherapy regimens. By this way,
individual analysis can be done to anticipate side effects of chemotherapy, especially hematologic toxicity.
Conflicts of interest
The authors affirm no conflict of interest in this
study
Acknowledgment
The authors highly appreciate the support from
Head of Surgical Oncology Department of Adam
Malik Hospital, Medan, dr. Emir Taris Pasaribu,
SpB(K)Onk; Head of Basic Medical Science
Postgraduate Program, Faculty of Medicine Universitas Sumatera Utara, dr. Yah Wardiyah Siregar, PhD, and the support from dr. Datten
Bangun, Msc, SpFK, Mardiah Nasution, Indra
Wahyudi, and Donny Nauphar.
REFERENCES
1. Thirumaran R, Prendergast GC, Gilman PB. Cytotoxic chemotherapy in clinical treatment of cancer. In: Prendergast GC, Jaffee EM, editors. Cancer immunotherapy: immune suppresion and tumor growth. USA: Elsevier Inc; 2007. p. 101–16.
2. Daniel D, Crawford J. Myelotoxicity from chemotherapy.
Semin Oncol. 2006 Nov;33(1):74–85.
3. Zhang J, Tian Q, Zhou SF. Clinical pharmacology of
cyclophosphamide and ifosfamide. Curr Drug Ther. 2006 Jan;1(1):55–84.
4. Tew KD, Manevich Y, Grek C, Xiong Y, Uys J, Townsend
DM. The role of glutatione S-Transferase P in signalling pathways and S-glutathionylation in cancer. Free Radic
Biol Med. 2011 July;51(2):299–313.
5. Zhong SL, Zhou SF, Chen X, Chan SY, Chan E, Ng KY, et al.
Relationship between genotype and enzyme activity of glutathione S-transferases M1 and P1 in Chinese. Eur J Pharm Sci. 2006 Jan;28(1–2):77–85.
6. Zhang BL, Sun T, Zhang BN, Zheng S, Lü N, Xu BH, et al.
Polymorphisms of GSTP1 is associated with differences of chemotherapy response and toxicity in breast cancer. Chin Med J. 2011 June;124(2):199–204.
7. Ge J, Tian A, Wang QS, Kong PZ, Yu Y, Li XQ, et al.
The GSTP1 105Val allele increases breast cancer risk and aggressiveness but enhances response to cyclophosphamide chemotherapy in North China. PLoS
One. 2013 June;8(6):e67589.
8. Tran A, Bournerias F, Le Beller C, Mir O, Rey E,
Pons G, et al. Serious haematological toxicity of cyclophosphamide in relation to CYP2B6, GSTA1 and GSTP1 polymorphisms. Br J Clin Pharmacol. 2008 Sep;65(2):279–80.
9. Ekhart C, Doodeman VD, Rodenhuis S, Smits PH,
Beijnen JH, Huitema AD. Influence of polymorphisms
of drug metabolizing enzymes (CYP2B6, CYP2C9,
cyclophosphamide and 4-hydroxycyclophosphamide.
Pharmacogenet Genomics. 2008 Feb;18(6):515–23.
10. Artini IGA, Astuti I, Purwono S, Aryandono T. The association between glutathione S-Transferase P1 polymorphism and myelotoxicity in breast cancer patients receiving cyclophosphamide-contained chemotherapeutic regimen. Indones J Cancer. 2011 Sep;5(4):1–7.
11. Zhong S, Huang M, Yang X, Liang L, Wang Y, Romkes M, et
al. Relationship of glutathione S-transferase genotypes with side-effects of pulsed cyclophosphamide therapy in patients with systemic lupus erythematosus. Br J Clin Pharmacol. 2006 July;62(4):457–72.
12. ctep.cancer.gov [Internet]. Cancer therapy evaluation program: common terminology criteria for adverse events ( CTCAE ). [update 2006 August 9; cited 20 Jul 2012]. Available from: http://ctep.cancer.gov/.
13. Keswara MA, Sudarsa IW, Golden N. The risk factor
of neutropenia on locally advanced breast cancer
patients treated with first cycle cyclophosphamide,
doxorubicine, 5- Fluorouracil chemotherapy at Sanglah General Hospital Denpasar, Bali-Indonesia. Bali Med J.
2012 Sep;1(3):116–20.
14. Han Y, Yu Z, Wen S, Zhang B, Cao X, Wang X. Prognostic
value of chemotherapy-induced neutropenia in early-stage breast cancer. Breast Cancer Res Treat. 2012
Oct;131(2):483–90.
15. Palappallil DS, Nair BL, Jayakumar KL, Puvathalil RT. Comparative study of the toxicity of
5-fluorouracil-cyclophosphamide versus
adriamycin-cyclophosphamide followed by paclitaxel in carcinoma
breast. Indian J Cancer. 2011 Jan;48(1):68–73.
16. Balmukhanov TS, Khanseitova AK, Nigmatova
VG, Ashirbekov EE, Talaeva SZ, Aitkhozhina NA.
Polymorphisms at GSTM1, GSTP1, GSTT1 detoxification genes loci and risk of breast cancer in Kazakhstan population. Sci Res. 2013 Oct;2:114–8.
17. Huang MY, Wang YH, Chen FM, Lee SC, Fang WY, Cheng TL, et al. Multiple genetic polymorphisms of GSTP1
313AG, MDR1 3435CC, and MTHFR 677CC highly
correlated with early relapse of breast cancer patients
in Taiwan. Ann Surg Oncol. 2008 Jan;15(3):872–80.
18. de Aguiar ES, Giacomazzi J, Schmidt AV, Bock H, Saraiva-Pereira ML, Schuler-Faccini L, et al. GSTM1, GSTT1, and GSTP1 polymorphisms, breast cancer risk factors and mammographic density in women submitted to breast cancer screening. Rev Bras Epidemiol. 2012;15(2):246–55.
19. Vivenza D, Feola M, Garrone O, Monteverde
M, Merlano M, Lo Nigro C. Role of the renin-angiotensin-aldosterone system and the glutathione S-transferase Mu, Pi and Theta gene polymorphisms in cardiotoxicity after anthracycline chemotherapy
for breast carcinoma. Int J Biol Markers. 2013 June;28(4):e336–47.
20. Ekhart C, Rodenhuis S, Smits PH, Beijnen JH, Huitema AD. An overview of the relations between polymorphisms in drug metabolising enzymes and drug transporters and survival after cancer drug treatment. Cancer Treat Rev.