Transcriptional control of plant genes responsive to pathogens
Paul J Rushton and Imre E Somssich
∗
Transcriptional activation of genes is a vital part of the plants defence system against pathogens.Cis-acting elements within the promoters of many of these genes have recently been defined and investigators have started to isolate their cognatetrans-acting factors. Some of these factors have counterparts in animals, whereas others are present only in plants, reflecting the fact that plants have developed a unique defence system.
Addresses
Max-Planck-Institut F ¨ur Zuchtungsforschung, Department of Biochemistry, Carl-von-Linn ´e-Weg 10, D-50829 K ¨oln, Germany ∗e-mail: [email protected]
Current Opinion in Plant Biology1998,1:311–315 http://biomednet.com/elecref/1369526600100311
Current Biology Ltd ISSN 1369-5266
Abbreviations
4CL 4-coumarate:CoA ligase
EREBP ethylene-responsive element binding protein
JA jasmonic acid
MRE MYB recognition element
PR pathogenesis-related
PAL phenylalanine ammonia-lyase
SA salicyclic acid
Introduction
Perception of a pathogen by a plant triggers rapid defence responses via a number of signal transduction pathways. A major target of signal transduction is the cell nucleus where the terminal signals lead to the transcriptional activation of numerous genes and consequently to the de novo synthesis of a variety of proteins and antimicrobial compounds [1,2•]. Studies with different plant pathology systems have revealed that the active response of plants to attempted microbial ingress is associated with dramatic reprogramming of cellular metabolism [2•]. Expression of a large array of genes whose products are involved both in diverse primary and secondary metabolic pathways are rapidly induced or strongly upregulated. These include genes encoding enzymes of the shikimate and the general phenylpropanoid pathways along with enzymes from subsequent branch pathways. Another group of genes, termed pathogenesis-related (PR) genes (comprised of classes 1–10), has often been found associated with the defence response [3]. The function of many PR proteins remains unknown but some are glucanases and chitinases. β-glucanases hydrolyse β-linked glucans, whereas chitinases degrade chitin, the major structural component of the cell walls of many fungi. Additionally, some plant chitinases also have lyzozymal activity. β-1,3 glucanase, in concert with chitinase have been shown to
degrade fungal cell walls and to inhibit fungal growth both
in vivoandin vitro[3].
The expression of pathogen-responsive genes shows both a temporal and spatial hierarchy. Some genes undergo a rapid localised activation at the site of infection, whereas others are more slowly activated either locally and/or systemically. Plant hormones, including salicylic acid (SA), ethylene and jasmonic acid (JA), have been implicated in acting as secondary signals following pathogen attack and enhanced expression of many pathogen-responsive genes can be induced by these plant hormones [4••].
Regulatory components of
pathogen-responsive genes
Differences in the expression patterns of pathogen-respon-sive genes are a result of the architecture of the promoters; pathogen-responsive promoters being dissimilar in the number, order and type of regulatory elements present. This places the promoters at the end of one or more signal transduction pathways as the plant responds to pathogen attack. Despite the differences in architecture, functional dissection of the promoter regions of some of these genes is starting to uncover common cis-acting elements and has provided us with the tools to identify distinct DNA-binding proteins required to modulate transcription. Interestingly, some of the cognate factors, for example EREBP and WRKY proteins, appear unique to plants whereas others such as bZIP and MYB proteins also have counterparts in animals.
The GCC Box and EREBPs
The GCC Box (AGCCGCC; Figure 1) is found in the promoter regions of many pathogen-responsive genes [5,6•]. Thiscis-acting element has been shown to function as an ethylene response element [5]. The biosynthesis of ethylene increases rapidly during plant–pathogen interactions and ethylene then acts through the GCC Box to induce the expression of pathogen-responsive genes [5,7••]. Interestingly, although similar elements also act as low temperature response elements and water deficit response elements, the GCC Box has yet to be found in the promoters of ethylene-regulated genes involved in some of the other ethylene responses, such as fruit ripening [8•,9]. It seems, therefore, that ethylene perception involves distinct regulatory mechanisms.
A number of proteins that bind to GCC Boxes have been isolated and found to be members of the EREBP (ethylene-responsive element binding protein) family of DNA-binding proteins [5,6•,7••,8•]. EREBPs contain a DNA-binding domain that is also present in the APETALA2 family of proteins [6•]. The homeotic gene
Figure 1
Current Opinion in Plant Biology MYB
MYB KAP-2 KAP-1
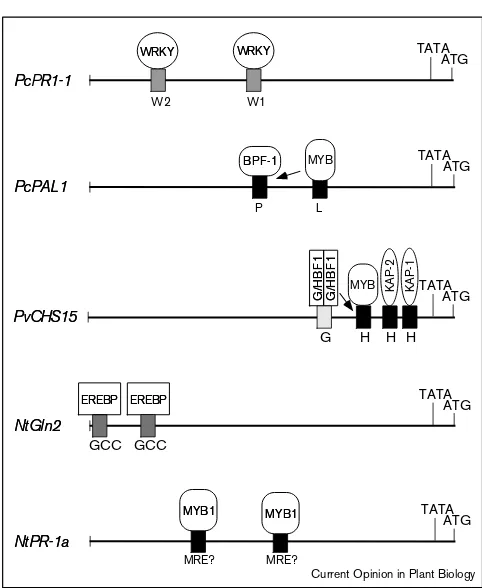
Schematic representation of pathogen responsivecis-acting elements (W-, P-, L-, G-, H-boxes, GCC-MRE-elements) and their cognate interacting factors (WRKY, BPF-1, MYB, G/HBF1, KAP1/2, EREBP). The promoter regions of thePetroselinum crispumPR1-1 (PcPR1-1) and phenylalanine ammonia-lyase 1 (PcPAL1), thePhaseolus vulgaris
chalcone synthase (PvCHS15), and theNicotiana tabacumglucanase
2 (NtGln2), and PR1 (NtPR-1a) genes are shown. The relative
positions of the interacting partners in relation to their respective TATA-boxes and the translation start sites (ATG) are given but are not drawn to scale. bZIP factors, such cs G/HBF1, are known to bind as dimers. Thus, a homodimer of G/HBF1 is indicated binding to the G-box although it cannot be excluded that G/HBF1 binds instead with an unknown second factor as a heterodimer to this element. Arrows point to additional elements which may be contacted by the respective factors as described in the text.
flower development in Arabidopsis thaliana[10]. The first EREBPs to be isolated were tobacco EREBP-1, -2, -3 and -4 [5]. All four bind specifically to GCC Boxes and are ethylene-inducible in leaves. More recently, CBF1, an EREBP from A. thaliana, was isolated by a yeast one-hybrid screen using a dehydration response element (GCCGAC) as the ‘bait’ DNA [8•]. This suggests that members of the EREBP family may play a wider role in the response to various stresses. In addition, CBF1 is capable of transactivation in yeast and this requires a functional cis-acting element [8•]. Further evidence for a role for EREBPs in plant defence has come from yeast two-hybrid experiments using the tomato resistance gene
Pto as a ‘bait’ protein [7••]. Pto confers resistance to
Pseudomonas syringae strains carrying the corresponding
avrPto avirulence genes. Pto is a protein kinase and was shown to interact with three proteins: Pti4, Pti5 and Pti
6. Pti4/5/6 are EREBPs and bind specifically to GCC boxes. In tobacco carrying a Pto transgene, expression of an EREBP gene was specifically enhanced upon infection with a bacterial strain harboring the avrPto avirulence gene. This suggests a direct link between a resistance gene and the specific activation of plant defence genes and implies that Pto confers resistance by activating a signalling pathway that leads through EREBPs to activation of PR genes containing GCC boxes.
W Boxes and WRKY proteins
There are two types of well studied W Box [11] elements, the first contains the hexamer sequence TTGACC whilst the second consists of the two tetramer half sites TGAC-Nx-GTCA. A W Box in the parsley PR-10 class gene, PR1-1, is the site of a fungal elicitor-inducible DNA-binding activity [12]. A similar activity also binds to a W Box in the promoter of the tobacco chitinase geneCHN50[13]. Experiments have shown not only that W Boxes are functional elicitor response elements in the parsleyPR1genes [11], the maize PR-1 class gene PRms
[14], and the tobacco class I chitinase geneCHN50[13,15] but also that they are sufficient to direct elicitor responsive expression in transient expression systems [11,14]. Other pathogen-responsive promoters contain W Box sequences within functional areas of the promoters. These include the potato glutathioneS-transferasegst1(prp1) [16], PR10 genes from asparagus (AoPR1 ) [17] and potato (PR-10a) [18], and the stilbene synthase gene,Vst1, from grapevine encoding an enzyme of phytoalexin biosynthesis [19]. Thus, W Boxes may be a general feature of a large subset of pathogen-responsive gene promoters.
elicitor induces rapid and transient changes in the mRNA levels of WRKY1, -2 and -3 [11]. Thus, WRKY proteins appear to be transcription factors that play a role during plant defence by binding to W Boxes in the promoters of pathogen-responsive genes.
MRE-like sequences and MYB factors
Another class of cis-acting elements is represented by Boxes P and L from the parsley phenylalanine ammonia-lyase (PAL) and 4-coumarate:CoA ligase (4CL) genes and the H-box from the bean chalcone synthase gene
Chs15 [23,24]. These elements fit the type II MYB consensus sequence A(A/C)C(A/T)A(A/C)C, suggesting that they are MYB recognition elements (MREs). Boxes P and L were initially identified within the parsley
PAL1 promoter as the sites of fungal elicitor-inducible DNA–protein interactions [25] and were subsequently shown to bind a MYB-like protein, BPF-1 [26,27•]. BPF-1 mRNA accumulates rapidly in elicitor-treated parsley cells suggesting that it participates in the plant defence response [26]. Boxes P and L from the parsleyPAL1,4CL1 and 4CL2 promoters can be specifically bound by MYB proteins [27•]. Furthermore, expression of the tobacco4CL
gene, along with other MRE-containing genes encoding enzymes of phenylpropanoid metabolism, was significantly repressed in transgenic tobacco plants overexpressing an Antirrhinum majus MYB protein [28•]. Additionally, a flower-specific MYB can activate transcription from the
A. thaliana gPAL2 promoter through similar elements [29]. Functional studies with the H-box indicated that it cannot function to a high level alone. Gain of function experiments, however, show that it is active in combination with a G-Box element (see below) in transgenic tobacco plants in establishing the characteristic tissue-specific pattern of expression and mutations in either the H-box or G-Box reduced the response to tobacco mosaic virus (TMV) infection [24,30].
Finally, the tobacco MYB1 gene is induced by TMV during the hypersensitive response and the development of systemic acquired resistance [31]. Application of SA induced expression ofMYB1within 15 minutes. Although the MYB1 protein can bind in vitro to a MYB consensus binding site found in the tobacco PR1-a promoter [31], it remains unclear whether this DNA element is of functional importance for PR1-a expression and whether it represents thein vivotarget site for MYB1.
G-Boxes,as-1-like elements and bZIP factors
G-Boxes (CACGTG) function during the regulation of diverse genes by environmental cues, such as abscisic acid (ABA), light, UV radiation and wounding, as well as pathogen signals [32]. G-Boxes are members of the family of ACGT-containing cis-acting elements and have been implicated in the expression of a number of genes during pathogen attack [33]. Often they function in concert with other cis-acting elements as in the case of the G-Box and H-box from the beanChs15promoter [24].
Another class of elements implicated in the plant defense response, consists of as-1-like elements. The as-1 (or
ocs) element (CTGACGTAAGGGATGACGCAC) was
initially identified in the 35S promoter of cauliflower mosaic virus and thenosandocspromoters ofAgrobacterium tumifaciens [34,35]. The as-1 element confers responsive-ness to several signals, including SA, auxin, jasmonates and hydrogen peroxide [4••]. Although the as-1 element is responsible for the activation of a number of pathogen genes, it is not clear whether functionalas-1-like elements are found widely in plant genes [34].
Both the G-Box and the as-1 or ocs element are bound by bZIP proteins, a well characterised class of transcription factors also found in animals where they regulate, both positively and negatively, various cellular processes. bZIP proteins appear to play important roles during the activation of a number of genes during plant defence. A bZIP protein from soybean binds to the G-Box in the bean Chs15 promoter [36•]. This protein, G/HBF-1, can also bind to the adjacent H-box. The significance of this relaxed binding specificity is unclear. Although the mRNA and protein levels of G/HBF-1 do not increase during the induction of its putative target genes, the protein itself is rapidly phosphorylated andin vitrophosphorylation enhances binding to one (H-box III) out of the three H-boxes present in theChs15 promoter. A serine kinase capable of phosphorylating G/HBF-1 was also identified and this kinase is rapidly and transiently stimulated in elicitor-treated cells [36•]. B ¨uttner and Singh [6•] showed that an ocs element-binding protein (OBF4) interacts with an EREBP suggesting that cross-coupling between EREBP and bZIP factors may be involved in the regulation of gene expression during the plant defence response.
Apart from being a binding site for bZIP proteins, the G-Box is also a potential basic helix-loop-helix bHLH protein binding site (CACNTG) [37]. bHLH factors are present in plants [38] but although MYB/bHLH interacting partners control flower pigmentation [39•], no role has yet been assigned to them during plant defence.
Additional regulatory elements and DNA-binding proteins
Salicylic acid (SA) is an important signal in plant defence [4••] and a SA-response element (SARE) has been identified in the tobacco PR2-d gene. A 76 bp fragment conferred a 20-fold induction by SA in transgenic tobacco plants and protein binding studies indicated that the core sequence of this SARE is TTCGACCTCC [40]. The nature of the binding factor is unknown.
that is identical to Box W3 from parsley suggesting that the region contains a W Box and, by implication, that PBF-1 and PBF-2 are WRKY proteins.
Two H-box binding activities, KAP-1 and KAP-2, have been purified from bean [42]. Elicitation with glutathione did not affect their total cellular activities but instead increased their specific activities in the nuclear fraction. KAP-1 and KAP-2 appear different from the other characterised H-box binding factor G/HBF but their identities remain unknown.
In some cases, promoter areas have been delineated that contain pathogen-responsivecis-acting elements that have yet to be clearly defined [19,43–45]. Sometimes these regions contain sequences similar to already characterised elements, but this is not always the case. Similarly, less well characterised proteins have also been implicated in the regulation of defence-related genes [46,47]. The future will certainly see additions to our list of regulatory elements and cognate transcription factors.
Conclusions
The past few years have marked a period of significant progress in defining both cis-acting elements within the regulatory regions of pathogen-responsive genes and the
trans-acting factors that bind them. Continuing efforts should add to this knowledge and also facilitate the isolation of additional signal transduction components upstream in these pathways. From a biotechnology standpoint, these discoveries may prove valuable in gene transfer strategies aimed at improving plant tolerance to pathogens. Looking to the future, the biggest challenge will be the establishment of causal in vivo relationships between the binding of distinct or overlapping groups of factors to specific elements and the regulation of target gene expression.
Acknowledgements
We thank all colleagues for providing preprints and unpublished data and Klaus Hahlbrock, Robert Cormack, Thomas Eulgem, Silke Robatzek and Bernd Weisshaar (all K ¨oln) for discussions and critical reading of the manuscript.
References and recommended reading
Papers of particular interest, published within the annual period of review, have been highlighted as:
• of special interest
•• of outstanding interest
1. Hammond-Kosack KE, Jones JDG:Resistance gene-dependent plant defense responses.Plant Cell1996,8:1773-1791.
•
2. Somssich IE, Hahlbrock K:Pathogen defence in plants — a paradigm of biological complexity.Trends Plant Sci1998,3 :86-90.
A short review focusing on the biochemical complexity of the plant defence response set in motion upon pathogen attack.
3. Kombrink E, Somssich IE:Pathogenesis-related proteins and plant defense.InThe Mycota Part A, Plant Relationships, Edited by Carroll G, Tudzynski P. Berlin: Springer-Verlag; 1997:107-128.
••
4. Yang Y, Shah J, Klessig DF:Signal perception and transduction in plant defense responses.Genes Dev1997,11:1621-1639.
An excellent review dealing with various aspects of plant defence including pathogen perception, signal transduction and activation of local and sys-temic responses. Genetic approaches in dissecting signalling pathways are also discussed.
5. Ohme-Takagi M, Shinshi H:Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element.
Plant Cell1995,7:173-182.
•
6. B ¨uttner M, Singh KB:Arabidopsis thalianaethylene-responsive element binding protein (AtEBP), an ethylene-inducible, GCC box DNA-binding protein interacts with an ocs element binding protein.Proc Natl Acad Sci USA1997,94:5961-5966.
Data is presented showing that interaction between EREBP and bZIP transcription factors can occur and that such cross-coupling may be impor-tant in regulating gene expression during the plant defence response.
••
7. Zhou J, Tang X, Martin GB:The Pto kinase conferring resistance to tomato bacterial speck disease interacts with proteins that bind acis-element of pathogenesis-related genes.EMBO J
1997,16:3207-3218.
The identification of three tomato EREBP-type proteins (Pti4/5/6) which in-teract with the Pto kinase is described, using the yeast two-hybrid system. Pti4/5/6 specifically recognized and bound GCC boxes that are present within the promoters of severalPRgenes. This work establishes, for the first time, a direct link between a plant resistance gene and activation of downstream defence genes.
•
8. Stockinger EJ, Gilmour SJ, Thomashow MF:Arabidopsis thaliana CBF1encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit.Proc Natl Acad Sci USA
1997,94:1035-1040.
The isolation anArabidopsisfactor, CBF1, is described which binds specifi-cally to DNA elements present in the promoters of many cold-regulated plant genes. The CBF1 protein contains an AP2 DNA-binding domain also present in EREBPs, suggesting that EREBP-like proteins may be involved in regulating diverse genes responsive to different stresses.
9. Deikman J:Molecular mechanisms of ethylene regulation of gene transcription.Physiol Plant1997,100:561-566.
10. Jofuku KD, den Boer BGW, van Montagu M, Okamuro JK:Control ofArabidopsisflower and seed development by the homeotic geneAPELATA2. Plant Cell1997,115:437-451.
11. Rushton PJ, Torres JT, Parniske M, Wernert P, Hahlbrock K, Somssich IE:Interaction of elicitor-induced DNA binding proteins with elicitor response elements in the promoters of parsley PR1 genes.EMBO J1996,15:5690-5700.
12. Meier I, Hahlbrock K, Somssich IE:Elicitor-inducible and constitutive in vivo DNA footprints indicate novelcis-acting elements in the promoter of a parsley gene encoding pathogenesis-related protein 1.Plant Cell1991,3:309-315. 13. Fukuda Y:Interaction of tobacco nuclear proteins with an
elicitor-responsive element in the promoter of a basic class I chitinase gene.Plant Mol Biol1997,34:81-87.
14. Ravent ´os D, Jensen AB, Rask M-B, Casacuberta JM, Mundy J, San Segundo B:A 20 bycis-acting element is both necessary and sufficient to mediate elicitor response of a maize PRms gene.
Plant J1995,7:147-156.
15. Fukuda Y, Shinshi H:Characterization of a novelcis-acting element that is responsive to a fungal elicitor in the promoter of a tobacco class I chitinase gene.Plant Mol Biol1994,
24:485-493.
16. Hahn K, Strittmatter G:Pathogen defence geneprp1-1
from potato encodes an auxin-responsive glutathioneS -transferease.Eur J Biochem1994,226:619-626. 17. Warner SAJ, Scott R, Draper J:Isolation of an asparagus
intracellularPRgene (AoPR1) wound-responsive promoter by the inverse polymerase chain reaction and its characterization in transgenic tobacco.Plant J1993,3:191-201.
18. Matton DP, Prescott G, Bertrand C, Camirand A, Brisson N:
Identification of cis-acting elements involved in the regulation of the 17 kDa pathogenesis-related geneSTH-2in potato.Plant
Mol Biol1993,22:279-291.
20. de Pater S, Greco V, Pham K, Memelink J, Kijne J:
Characterization of a zinc-dependent transcriptional activator fromArabidopsis.Nucleic Acids Res1996,24:4624-4631. 21. Rushton PJ, Macdonald H, Huttly AK, Lazarus CM, Hooley R:
Members of a new family of DNA-binding proteins bind to a conservativecis-element in the promoters ofα-Amy2genes.
Plant Mol Biol1995,29:691-702.
22. Mackay JP, Crossley M:Zinc fingers are sticking together.
Trends Biochem Sci1998,23:1-4.
23. Logemann E, Parniske M, Hahlbrock K:Modes of expression and common structural features of the complete phenylalanine ammonia-lyase gene family in parsley.Proc Natl Acad Sci USA
1995,92:5905-5909.
24. Faktor O, Kooter JM, Loake GJ, Dixon RA, Lamb CJ:Differential utilization of regulatorycis-elements for stress-induced and tissue-specific activity of a French bean chalcone synthase promoter.Plant Sci1997,124:175-182.
25. Lois R, Dietrich A, Hahlbrock K, Schulz W:A phenylalanine ammonia-lyase gene from parsley: structure, regulation and identification of elicitor and light responsivecis-acting elements.EMBO J1989,8:1641-1648.
26. da Costa e Silva O, Klein L, Schmelzer E, Trezzini GF,
Hahlbrock K:BPF-1, a pathogen-induced DNA-binding protein involved in the plant defense response.Plant J1993,4 :125-135.
•
27. Feldbr ¨ugge M, Sprenger M, Hahlbrock K, Weisshaar B:PcMYB1, a novel plant protein containing a DNA-binding domain with one MYB repeat, interactsin vivowith a light-regulatory promoter unit.Plant J1997,11:1079-1093.
The isolation of a parsley MYB-like factor and demonstration thatcis-acting elements such as the P- and L- boxes, common to several phenylpropanoid gene promoters, are MYB recognition elements (MREs).
•
28. Tamagnone L, Merida A, Parr A, Mackay S, Culianez-Macia FA, Roberts K, Martin C:The AmMYB308 and AmMYB330 transcription factors from Antirrhinum regulate phenylpropanoid and lignin biosynthesis in transgenic tobacco.
Plant Cell1998,10:135-154.
The demonstration that overexpression of twoAntirrhinumMYB proteins in tobacco plants specifically repressed phenolic acid metabolism and lignin biosynthesis probably by binding to their normal target genes.
29. Sablowski RWM, Moyano E, Culianez-Macia FA, Schuch W, Martin C, Bevan M:A flower-specific Myb protein activates transcription of phenylpropanoid biosynthetic genes.EMBO J
1994,13:128-137.
30. Faktor O, Loake G, Dixon RA, Lamb CJ:The G-box and H-box in a 39 bp region of a French bean chalcone synthase promoter constitutes a tissue-specific regulatory element.Plant J1997,
11:1105-1113.
31. Yang Y, Klessig DF:Isolation and characterization of a tobacco mosaic virus-induciblemyboncogene homolog from tobacco.
Proc Natl Acad Sci USA1996,93:14972-14977.
32. Menkens AE, Schindler U, Cashmore AR:The G-box: a ubiquitous regulatory DNA element in plants bound by the GBF family of bZIP proteins.Trends Biochem Sci1995,20 :506-510.
33. Kim S-R, Choi J-L, Costa MA, An G:Identification of G-box sequence as an essential element for methyl jasmonate
response of potato proteinase inhibitor II promoter.Plant
Physiol1992,99:627-631.
34. Ellis JG, Tokuhisa JG, Llewellyn DJ, Bouchez D, Singh K, Dennis ES, Peacock WS:Does theocs-element occur as a functional component of the promoters of plant genes?Plant J1993,
4:433-443.
35. Lam E, Benfey PN, Gilmartin PM, Fang R-X, Chua N-H: Site-specific mutations alterin vitrofactor binding and change promoter expression pattern in transgenic plants.Proc Natl
Acad Sci USA1989,86:7890-7894.
•
36. Dr ¨oge-Laser W, Kaiser A, Lindsay WP, Halkier BA, Loake GJ, Doerner P, Dixon RA, Lamb C:Rapid stimulation of a soybean protein-serine kinase that phosphorylates a novel bZIP DNA-binding protein, G/HBF-1, during the induction of early transcription-dependent defenses.EMBO J1997,16:726-738. Isolated a soybean bZIP factor,G/HBF-1, that binds to both the G- and H-box. In addition, a cytosolic kinase was identified whose activity increases rapidly upon elicitation of soybean cells and enhances binding of G/HBF-1 to its DNA target.
37. Kawagoe Y, Murai N:A novel basic region/helix-loop-helix protein binds to a G-box motif CACGTG of the bean storage protein beta-phaseolin gene.Plant Sci1996,116:47-57. 38. de Pater S, Pham K, Memelink J, Kijne J:RAP-1 is anArabidopsis
MYC-like R protein homologue, that binds to G-box sequence motifs.Plant Mol Biol1997,34:169-174.
•
39. Martin C, Paz-Ares J:MYB transcription factors in plants.Trends
Genet1997,13:67-73.
A nice review on the role of plant MYB transcription factors in controlling secondary metabolism, cell morphology, cellular proliferation and plant re-sponses to hormones.
40. Shah J, Klessig DF:Identification of a salicylic acid-responsive element in the promoter of the tobacco pathogenesis-related rn-1,3-glucanase gene,PR-2d.Plant J1996,10:1089-1101. 41. Despr ´es C, Subramaniam R, Matton DP, Brisson N:The activation
of the potatoPR-10agene requires the phophorylation of the nuclear factor PBF-1.Plant Cell1995,7:589-598.
42. Yu LM, Lamb CJ, Dixon RA:Purification and biochemical characterization of proteins which bind to the H-boxcis -element implicated in transcriptional activation of plant defense genes.Plant J1993,3:805-816.
43. Rouster J, Leah R, Mundy J, Cameron-Mills V:Identification of a methyl jasmonate-responsive region in the promoter of a lipoxygenase 1 gene expressed in barley grain.Plant J1997,
11:513-523.
44. Grimmig B, Matern U:Structure of the parsley caffeoyl-CoAO -methyltransferase gene, harbouring a novel elicitor responsive
cis-acting element.Plant Mol Biol1997,33:323-341. 45. Yin S, Mei L, Newman J, Back K, Chappell J:Regulation of
sesquiterpene cyclase gene expression. Characterization of an elicitor- and pathogen-inducible promoter.Plant Physiol1997,
115:437-451.
46. Brandt J, Thordal-Christensen H, Vad K, Gregersen PL, Collinge DB:A pathogen-induced gene of barley encodes a protein showing high similarity to a protein kinase regulator.Plant J
1992,2:815-820.