Cosmid vectors such as pOCA18 (Ref. 1) and pLZO3 (Ref. 2) have been developed that can transform plants. However, the low cloning capacity of these vectors limits their
useful-ness. Recently, two research groups have developed vectors for the transformation of large DNA fragments into plants3,4. These vectors are designed to have the
characteris-tics of both bacterial artificial chromosomes (BACs) and binary vectors forAgrobacterium -mediated transformation. Therefore, large DNA fragments cloned in the T-DNA region can be transferred into a plant genome by Agrobacterium. Indeed, many genomic-DNA fragments (<150 kb) have been introduced successfully into several plant species by these vectors4,5.
This transformation technology has con-tributed to the map-based cloning (positional cloning) of novel genes defined by mutation from Arabidopsis4,6. It should facilitate the iso-lation of dominant alleles of agronomic impor-tance. Furthermore, the technology will be useful in plant biotechnology, especially for situations that require the simultaneous introduction of several genes, such as those encoding the enzymes of a metabolic pathway.
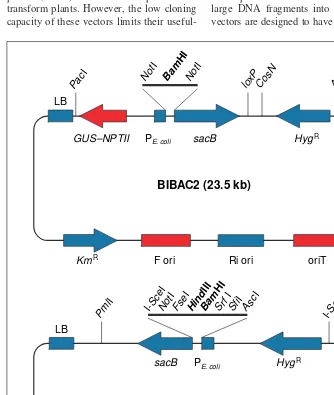
Structure and function of the BAC vectors The binary bacterial artificial chromosome (BIBAC) vector BIBAC2 (Ref. 3) and the transformation-competent bacterial artificial chromosome (TAC) vector pYLTAC7 (Ref. 4) have similar components (Fig. 1):
• Positive-selection markers active in E. coli (the sacBgene).
• Elements for the stable maintenance of foreign DNA in either E. coli(the F factor or P1 phage replication origin) or Agrobacterium tumefaciens(the Ri repli-cation origin of Agrobacterium rhizogenes). • cis sequences that are needed for Agro-bacterium-mediated gene transfer into plants (left and right border sequences).
• A selectable marker active in plants (hygromycin phosphotransferase). In addition to these frameworks, each vector also has unique components that allow clones to be handled or transgenes to be analyzed, but alternative methods can be used that avoid the need for most of these functions (Box 1). Protocols for handling the vectors and clones are available on the WWW (Box 2).
Transforming plants with BAC clones carrying large DNA fragments
A 150 kb human DNA fragment was trans-ferred into tobacco using the BIBAC vector system5. Most of the transgenic plants pro-duced (63%) are thought to have the entire 150 kb insert. Whole inserts (40–80 kb of genomic DNA) have also been successfully transferred into the Arabidopsisgenome using the TAC vector4. High molecular weight frag-ments introduced into plant genomes were inherited faithfully by progeny without further changes4,5. These results show that the trans-formation-competent BAC vectors work well for T-DNA transfer of, at least, 150 kb. However, no systematic studies on the factors that contribute to the complete transfer of large T-DNAs have yet been undertaken.
354
trends in plant science
Update
August 2000, Vol. 5, No. 8
Agrobacterium-mediated plant
transformation with large DNA
fragments
Fig. 1.The functional components of the binary bacterial artificial chromosome (BIBAC) vector BIBAC2 (Ref. 3) and the transformation-competent bacterial artificial chromosome (TAC) vec-tor pYLTAC7 (Ref. 4) (not to scale). The components that are given the same name in both the vectors (blue) have the same function but are not necessarily identical in sequence. The compo-nents that are unique to BIBAC2 or pYLTAC7 are red. Cutting sites for various restriction enzymes are indicated. The BamHI or HindIII sites are used for cloning insert DNA. The entire sequence of the vector pYLTAC7 is available from GenBank (Accession no. AB020028). Abbreviations: cosN, lcosN site; F ori, the F factor replication origin; GUS–NPTII, b-glu-curonidase and neomycin phosphotransferase II fusion; HygR
, hygromycin resistance gene; KmR , kanamycin resistance gene (NPT1); LB, left border sequence; loxP, P1 loxP site; PE. coli, a
syn-thetic E. colipromoter; RB, right border seqeunce; Ri ori, the replication origin of the Ri plasmid of Agrobacterium rhizogenes; sacB, the sacB gene of Bacillus amyloliquifaciens.
Trends in Plant Science
sacB HygR
RB LB
LB RB
GUS–NPTII
KmR
Km R
P1 replicon
P1 lytic replicon Ri ori
F ori Ri ori oriT
PE. coli sacB HygR
Pac I
Not I
loxPCosN Asc
I Not
I
Bam
HI
BIBAC2 (23.5 kb)
pYLTAC7 (22.9 kb) PE. coli Pml
I
I-Sce I
Not I
Fse I
Srf I
SfiI Asc I
Hind
III
Bam
HI
I-Sce I
The efficiency of Arabidopsis transfor-mation with a TAC clone with an 80 kb insert was 3.4 (482 transgenic plants from 142 plants inoculated by vacuum infiltration), and that with the TAC vector alone (4.4 kb T-DNA only) was 5.4 (688 transgenic plants from 127 treated plants)4. This indicates that there is a moderate dependency of the transformation efficiency on T-DNA length at least up to 80 kb. Tobacco transformation with low mol-ecular weight BIBAC T-DNAs produced about two transgenic plants per leaf strip treated3. However, the efficiency with the BIBAC clone carrying the 150 kb DNA using A. tumefaciens UIA143 was only 0.12 (33 transgenic plants from 272 inoculated leaf strips)5. In this experiment, the use of the viru-lence helper plasmids3, which overproduce either VirG or VirG and VirE in A. tumefaciens, was crucial for producing transgenic plants. Further study is needed to produce highly effi-cient transformation of large DNA fragments (~150 kb).
Various A. tumefaciensstrains can be trans-formed successfully with the transformation-competent BAC vectors. Several TAC clones carrying the ~80 kb inserts have been intro-duced into Arabidopsisusing A. tumefaciens strains C58C1(MP90), C58C1(GV2260) or EHA105 by a vacuum infiltration4. Standard root-transformation protocols also work well for Arabidopsis transformation using TAC clones6. The BIBAC vectors were introduced into a wide variety of A. tumefaciensstrains (EHA105, LBA4404, GV3101, MOG101, UIA143pMP90 and UIA143pMOG101) and all of the strains produced transgenic tobacco plants3. The A. tumefaciensstrains that are deficient in recAfunction (required for recom-bination), such as the UIA143 series (Ref. 7) and AGL1 (Ref. 2), might be useful for the sta-ble maintenance of some DNA fragments that contain repetitive sequences.
Map-based cloning using the BACs Complex biological processes such as the regu-lation of development and gene-expression cas-cades have been analyzed by molecular genetics. The many sets of DNA markers and genomic DNA libraries that have been prepared from various plant species help in the map-based cloning of novel genes. However, the major drawback of map-based cloning, is the difficulty of narrowing down the field of candidate clones to a manageable number for complementation testing.
The accuracy of the genetic distances between the target gene and the nearest DNA markers (defined by recombination frequency) depends on the number of progeny (usually F2 plants) tested. If mutations exhibit incomplete penetration, accuracy also depends on the pre-cision of scoring the mutant phenotype. About 1000 progeny are therefore required for
fine-scale mapping of a mutant locus. If the target locus is present in a chromosomal region that exhibits a low recombination frequency8, or a low level of DNA polymorphism, positional cloning becomes impractical.
The ability to transform a mutant plant with large fragments of genomic DNA that encom-pass the locus of the corresponding wild-type allele would overcome the principal drawback of current positional-cloning procedures. Therefore, transformation-competent BAC clones might facilitate map-based cloning. Once a large DNA fragment containing the target gene is isolated, DNA manipulations
(e.g. creating a nested deletion series from the fragment) and subsequent complementation test could identify the gene easily.
Genomic DNA libraries have been con-structed using transformation-competent BAC vectors from several plant species. BIBAC genomic DNA libraries have been constructed from two tomato species: Lycopersicon esculentum cv. Mogeor and Lycopersicon pennelliiLA716 (Ref. 9). L. esculentumcv. Mogeor contains the wild-type allele for most major morphological genes and a wide spectrum of disease-resistance genes, and L. pennellii LA716 contains several important quantitative
355
trends in plant science
Update
August 2000, Vol. 5, No. 8 Box 1. Unique characteristics of BIBAC and TAC vectors
In addition to the basic components of the binary bacterial artificial chromosome (BIBAC) vector BIBAC2 and the transformation-competent bacterial artificial chromosomes (TAC)
vector pYLTAC7, they have other useful features3,4. However, there are alternative methods that can be used to avoid the need for most, but not all, of these features.
• The RK2 oriTsequence of BIBAC2 (for triparental mating): electroporation is also a good method for delivering plasmids to Agrobacterium tumefaciens.
• T7 and SP6 RNA polymerase promoters of BIBACs (for generating RNA probes from the insert): thermal asymmetric interlaced-PCR (TAIL-PCR) works well for generating probes from TAC clone ends23,24. (This should also be applicable to the BIBAC clones.)
• The gene that encodes a bifunctional fusion peptide of b-glucuronidase and neomycin phos-photransferase II adjacent to the left border of BIBACs [selection for this marker adjacent might be used to identify transgenic plants that are likely to carry the entire T-DNA (Ref. 3)]: PCR amplification of other sequences adjacent to the left border (e.g. sacB) can be used for the same purpose after selecting hygromycin-resistant plants.
• The P1 lytic replicon of pYLTAC7 (overcomes the disadvantage of low yield during DNA preparation): in most cases, the yield of BAC-clone DNA is not a problem.
• Two I-SceI sites of pYLTAC7 [for Southern blot analysis of the integrity of large transgenes – I-SceI recognizes an 18-bp sequence and therefore most plants should not contain a I-SceI site, meaning that the entire insert sequence (including the HPTtag) can be released from the plant genome by I-SceI digestion and be detected by Southern analysis with an HPTprobe]: the rare-cutting enzymes NotI, PacI, AscI and Crerecombinase for loxP and l-terminase for
cosNcan be used for the same purpose if the insert does not contain these sites.
• The unique HindIII and BamHI sites of pYLTAC7 for library construction (the efficient ligation of HindIII cohesive ends ensures that large-scale genomic DNA libraries can be pre-pared easily using HindIII): genomic DNA libraries of wheat have been constructed using a modified TAC vector (Y-G. Liu and Y. Ogihara, unpublished). One of the libraries that con-tains 106independent clones with an average insert size of 60 kb.
Box 2. Protocols for handling and analyzing transformation-competent BAC vectors and clones
http://www.research.cornell.edu/biotech/BIBAC/bibachomepage/
Includes protocols for binary bacterial artificial chromosome (BIBAC) plasmid preparation from E. coliand Agrobacterium, the electroporation of BIBAC clones into bacteria and the con-struction of tomato genomic DNA BIBAC libraries, as well as other useful tips. The site also provides the information about the availability of these materials.
http://www.kazusa.or.jp/en/plant/TAC/
Provides protocols for transformation-competent bacterial artificial chromosome (TAC) plas-mid preparation and information about the construction of the Arabidopsis genomic-DNA library. The TAC vector is available from the Plant Cell Bank of the RIKEN Gene Bank (Tsukuba, Japan; e-mail [email protected]).
http://www.kazusa.or.jp/kaos/
Provides the sequences of the TAC clones used for genome sequencing of Arabidopsis
chromosomes III and V (Ref. 25). http://hbz.tamu.edu/
trait loci (QTLs). Rice and wheat genomic-DNA libraries have been constructed using a modified TAC vector (Y-G. Liu, unpublished). A TAC-based Arabidopsis (Columbia accession) genomic DNA library has also been constructed in which each plasmid contains a DNA insert of ~80 kb (Y-G. Liu and D. Shibata, http://www.kazusa.or.jp/en/plant/TAC/).
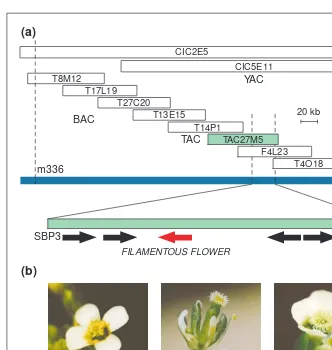
This TAC library has been used for the map-based cloning of novel genes. Several TAC clones were isolated covering the region containing the locus of the gravitropic mutant sgr1and then the sgr1phenotype was com-plemented with two TAC clones carrying genomic fragments of 75 kb or 80 kb (Ref. 4). The FILAMENTOUS FLOWERgene, which is important in the determination of meristem and organ identity in Arabidopsis, was also isolated with the use of the TAC cloning sys-tem6(Fig. 2). This gene encodes a protein with
a zinc finger and high-mobility-group-related (HMG-related) domains.
The CKH1 gene, which is involved in cytokinin signaling10, was isolated using a TAC clone (T. Kakimoto, pers. commun.). A temperature-sensitive mutation of Arabidopsis that has swollen roots at the non-permissive temperature was isolated using TAC clones (S. Sato et al., unpublished). Together, these results show that TAC-based cloning is a powerful way to isolate novel genes defined by mutation. Given that the Arabidopsis gen-ome will be completely sequenced before the end of the year 2000 (Ref. 11), sequencing the insert ends of all TAC clones of the Arabidopsislibrary is currently under way in order to construct a TAC-based physical map of the genome (D. Shibata, unpublished). Such a map will greatly increase the rate of positional cloning of Arabidopsisgenes.
Perspectives
The vector systems for plant transformation with large DNA fragments should prove to be useful for the positional cloning of previously unidentified genes from crops. Many genes of agronomic importance have been mapped to specific chromosomes of crop species12. For most crops, it is impractical to produce a satu-rated population of T-DNA transgenic lines similar to those generated for Arabidopsis13. Crop genes have been tagged and isolated with the use of transposons14and retrotrans-posons15. However, the tagging approach has severe limitations because of the null function of most tagged alleles and also because appro-priate transposons suitable for gene isolation are not available in most crops.
The positional cloning of genes that have been mapped to crop chromosomes is therefore desirable. Even in crops such as rice16, for which multiple DNA markers are available, it remains difficult to narrow down the chromosomal region of interest to a manageable size for gene identification. However, complementation test-ing with large genomic-DNA fragments is an alternative approach for the efficient identifica-tion of target genes if the crop of interest can be transformed by Agrobacterium. The construc-tion of crop genomic-DNA libraries using trans-formation-competent BAC vectors such as the BIBAC and TAC vectors should accelerate the positional cloning of novel genes from these species.
A potential problem associated with Agrobacterium-mediated transformation is the stability of large DNA fragments in A. tumefaciensstrains. Repetitive sequences, which are ubiquitous components of nuclear genomes in many species across the plant kingdom17, might not be maintained in bac-teria because the homologous-recombination system can remove part of such sequences. A. tumefaciens strains that are deficient in recAfunction [such as UIA143 (Ref. 7) and AGL1 (Ref. 2)] might help to overcome some of these problems. However, as homologous recombination is not the only factor that de-stabilizes foreign DNA in bacteria18, some plant DNA sequences could not be maintained stably even in recA-mutant strains.
An alternative way to transfer large DNA fragments into plants without passing through A. tumefacienscells is particle-bombardment technology. It was recently shown that large DNA fragments (<230 kb) cloned in the pBACwich vector for site-specific recombina-tion were transferred successfully to the tobacco genome by particle bombardment19. TAC or BIBAC clones might be applicable for trans-formation with particle bombardment. The technology might help in the transformation of crops that are resistant to Agrobacterium -medi-ated transformation. Electroporation should also be examined for large-DNA-fragment transfer.
356
trends in plant science
Update
August 2000, Vol. 5, No. 8
Fig. 2.Isolation of the FILAMENTOUS FLOWERgene using a transformation-competent bacterial artificial chromosome (TAC) clone. (a) Physical map of the FILAMENTOUS FLOWER region. The mutation locus was initially mapped between two markers, m336 and GBF3, which are present in yeast artificial chromosome clone CIC2E5. This region was then covered with several bacterial artificial chromosome clones and a TAC clone, TAC27M5 (Ref. 6). (b) Complementation of the mutation by the TAC clone. Fine mapping showed that the mutated gene was located between the markers SBP3 and ML. The FILAMENTOUS FLOWER
gene (red) was identified among five open-reading-frames (arrows) of this region6
. Courtesy of Shinichiro Sawa, Tokyo Metropolitan University, Japan and Kiyotaka Okada, Kyoto University, Japan.
Trends in Plant Science
ML SBP3
T8M12 T17L19
T27C20 T13E15
T14P1
TAC27M5 CIC2E5
CIC5E11
TAC BAC
YAC (a)
(b)
FILAMENTOUS FLOWER
Wild type filamentous
flower mutant
Mutant complemented with TAC27M5
GBF3 m336
F4L23 20 kb
The introduction of large genomic-DNA fragments into crops will be especially impor-tant for the isolation of genes corresponding to QTLs. Because QTLs control such important agronomic properties as crop yield, disease resistance and stress tolerance, the cloning of the corresponding genes is of great economic importance20. Several QTLs have been mapped to crop chromosomes by using DNA markers20 but the map positions are only approximate and near-isogenic lines have therefore been pre-pared to narrow down the chromosomal regions that contain the QTLs (Ref. 21). The prepara-tion of near-isogenic lines will require at least several years of work, even if DNA-marker-assisted selection procedures are applied. Moreover, after isolating such lines, the identi-fied region containing a QTL remains, in gen-eral, too large to allow the isolation of the corresponding gene. Representing the entire genome of a crop with large DNA fragments in transformation-competent vectors should facili-tate the isolation of QTL genes.
Techniques for the stable introduction of multiple genes into crops or trees are desirable for future biotechnology. Many genes of agri-cultural or industrial importance have been iso-lated. The simultaneous introduction of independent gene cassettes into rice by particle bombardment has been described22, but these genes were integrated into a few loci of chro-mosomes by unknown mechanisms. Analysis of 125 transgenic rice plants by PCR indicated that three such lines harbored up to 13 foreign genes. Introducing large DNA fragments con-taining multiple genes into plants is an alterna-tive approach, one advantage of which is that only a limited number of transgenic plants is required to analyze gene expression (an impor-tant feature for crops, such as soybean, that are resistant to transformation). Precise control of the expression of each gene on a single DNA fragment is feasible. However, because the introduction of many DNA fragments into a vector is a time-consuming process, it will be important to develop convenient protocols for such ligation of multiple DNA fragments.
References
01Olszewski, N. and Ausbel, F.M. (1988) Specialized binary vector for plant transformation: expression of
the Arabidopsis thaliana AHASgene in Nicotiana
tabacum. Nucleic Acids Res. 16, 10765–10782
02Lazo, G.R. et al.(1991) A DNA transformation-competent Arabidopsisgenomic library in
Agrobacterium. Bio/Technology9, 963–967
03Hamilton, C.M. (1997) A binary-BAC system for plant transformation with high-molecular-weight DNA. Gene200, 107–116
04Liu, Y-G. et al. (1999) Complementation of plant mutants with large genomic DNA fragments by a transformation-competent artificial chromosome vector accelerates positional cloning. Proc. Natl.
Acad. Sci. U. S. A. 96, 6535–6540
05Hamilton, C.M. et al. (1996) Stable transfer of intact high molecular weight DNA into plant chromosomes. Proc. Natl. Acad. Sci. U. S. A.93, 9975–9979
06Sawa, S. et al. (1999) FILAMENTOUS
FLOWER, a meristem and organ identity gene of
Arabidopsis, encodes a protein with a zinc finger
and HMG-related domains. Genes Dev. 13, 1079–1088 (erratum 13, 2337)
07Farrand, S.K. et al.(1989) Construction of an
Agrobacterium tumefaciensC58 recA mutant.
J. Bacteriol.171, 5314–5321
08Schmidt, R. et al.(1995) Physical map and organization of Arabidopsis thaliana
chromosome 4. Science 270, 480–483
09Hamilton, C.M. et al. (1999) Construction of tomato genomic DNA libraries in a binary-BAC (BIBAC) vector. Plant J.18, 223–229
10 Kubo, M. and Kakimoto, T. The
CYTOKININ-HYPERSENSITIVEgenes of Arabidopsis
negatively regulate the cytokinin-signaling pathway for cell division and chloroplast development. Plant J.(in press)
11 Meyerowitz, E.M. (1999) Today we have naming of parts. Nature402, 731–732
12 Gale, M.D. and Devos, K.M. (1998) Plant comparative genetics after 10 years. Science282, 656–659
13 Parinov, S. and Sundaresan, V. (2000) Functional genomics in Arabidopsis: large-scale insertional mutagenesis complements the genome sequencing project. Curr. Opin. Biotechnol.11, 157–161
14 Walbot, V. (2000) Saturation mutagenesis using maize transposons. Curr. Opin. Plant Biol.3, 103–107
15 Kumar, A. and Bennetzen, J.L. (1999) Plant retrotransposons. Annu. Rev. Genet.33, 479–532
16 Nagamura, Y. et al.(1997) Rice molecular genetic map using RLFPs and its applications.
Plant Mol. Biol.35, 79–87
17 Schmidt, T. (1999) LINEs, SINEs and repetitive DNA: non-LTR retrotransposons in plant
genomes. Plant Mol. Biol.40, 903–910
18Harris, T.J. and Emtage, J.S. (1986) Expression of heterologous genes in E. coli. Microbiol. Sci.
3, 28–31
19Choi, S. et al.(2000) A new approach for the identification and cloning of genes: the pBACwich system using Cre/loxsite-specific recombination. Nucleic Acids Res.28, 19e
20Paterson, A.H. (1995) Molecular dissection of quantitative traits: progress and prospects.
Genome Res. 5, 321–333
21Yano, M. and Sasaki, T. (1997) Genetic and molecular dissection of quantitative traits in rice.
Plant Mol. Biol.35, 145–153
22Chen, L. et al. (1998) Expression and inheritance of multiple transgenes in rice plants. Nat.
Biotechnol.16, 1060–1064
23Liu, Y-G. and Whittier, R.F. (1995) Thermal asymmetric interlaced PCR: automatable amplification and sequencing of insert end fragments from P1 and YAC clones for chromosome walking. Genomics25, 674–681
24Liu, Y-G. and Huang, N. (1998) Efficient amplification of insert end sequences from bacterial artificial chromosome clones by thermal asymmetric interlaced PCR. Plant Mol. Biol. Rep.
16, 175–181
25Sato, S. et al. (1998) A physical map of
Arabidopsis thalianachromosome 3 represented
by two contigs of CIC YAC, P1, TAC and BAC clones.DNA Res.5, 163–168
Daisuke Shibata*
Kazusa DNA Research Institute, Yana 1532-3, Kisarazu, Chiba 292-0812, Japan.
Yao-Guang Liu
South China Agricultural University, Guangzhou 510642, People’s Republic of China.
*Author for correspondence
(tel 181 438 52 3906; fax 181 438 52 3948;
e-mail [email protected])
357
trends in plant science
Update
The review article by Jill. M.Stevenson, Imara Y. Perera, Ingo Heilmann, Staffan Persson and Wendy F. Boss (2000) Inositol signaling and plant growth. Trends Plant Sci. 5, 252–258 in the June 2000 issue of Trends in Plant Science contained an error. In Figure 1, the structure of phosphatidylinositol (4,5) bisphosphate [PtdIns(4,5)P2] was depicted incor-rectly. A corrected version of this portion of the figure is printed here. We apologize to the authors and our