GENOME-WIDE ASSOCIATION STUDY FOR NATURAL
VARIATION IN ENZYMATIC ACTIVITIES AND MOTILITY
TRAITS IN Xanthomonas campestris pv. campestris
NANI MARYANI
GRADUATE SCHOOL
BOGOR AGRICULTURAL UNIVERSITY
BOGOR
DECLARATION
I declare that this thesis entitled “Genome-Wide Association Study for Natural Variation in Enzymatic Activities and Motility Traits in Xanthomonas campestris pv. campestris” is a report of research work carried out by me through the guidance of my academic supervisors. This thesis has not been submitted in any form for any college except in INSA (Institute Nationale des Science Appliquées) Toulouse, France, part of the Double Degree Indonesia France Program collaborate with Bogor Agricultural University. Information obtained from published or unpublished work of others and help received during laboratory and field work have been acknowledged.
Bogor, October 2012
ABSTRACT
NANI MARYANI. Genome-Wide Association Study for Natural Variation in Enzymatic Activities and Motility Traits in Xanthomonas campestris pv. campestris. Under supervision of MATTHIEU ARLAT, LISDAR I SUDIRMAN, GAYUH RAHAYU.
Genome wide association studies are commonly used to study complex traits involved in human disease and in plant development. This research suggest that genome wide association is also feasible to study natural variation in bacterial pathogen like Xanthomonas campestris pv. campestris (Xcc). Studies on 50 Xcc strains were done by phenotyping four enzymatic activities and two types of motilities with genomic data available, AFLP and SNPs markers. The results showed that the sample size affects the number of AFLP markers detected were identified some genes that correspond to significant coding SNPs for amylase and endoglucanase activities. Significant markers identified in this association might be a candidate gene for those traits. It is necessary to follow-up these studies to confirm this hypothesis.
SUMMARY
NANI MARYANI. Genome-Wide Association Study for Natural Variation in Enzymatic Activities and Motility Traits in Xanthomonas campestris pv. campestris. Under supervision of MATTHIEU ARLAT, LISDAR I SUDIRMAN, GAYUH RAHAYU.
Genome-wide association studies (GWAS) has emerged as a promising tool to reveal the genetic basis of organisms. The goal of GWAS is to look at the join distribution between phenotype and genotype. With GWAS researcherss seek to identify regions of the genome where individuals that are phenotypically similar are also unusually closely related. This is the first GWAS research in bacteria. Xanthomonas campestris pv. campestris (Xcc) was chosen as a model bacteria since genomic data required for such studies are available. Xcc is the causal agent of black rot disease of cruciferous plants. This bacteria has been used as a model to study plant-pathogen interactions. Pathogenic species and pathovars show a high degree of host plant specificity and many also exhibit tissue specificity. Strains of this pathogen also show different abilities to cause disease on host plants. These characteristics give more attention to study both phenotypic and genotypic variations in this pathogen. The objectives of this study are to study phenotype and genotype variations among 50 strains of Xcc and to identify new markers associated with phenotypic variation.
Two virulence traits of Xcc were first studied i.e. their extracellular enzymes and motility then performed association studies using GWAS approach. Two different sets: neutral AFLP markers (Amplified Fragment Length Polymorphism) and SNPs (Single Nucleotide Polymorphisms) were used. Radial diffusion assays method in agar plates containing substrate was used for phenotyping four kinds of extracellular enzymes amylase, endoglucanase, polygalacturonase, and protease. Swarming and swimming motility was also measured with the diameter of spreading bacteria in MOKA agar medium. To verify that the strains were not inverted during the experiments, at the end of one replicate of protease activity, all the strains were verified by PCR using four couple of primers specific to xopAL2, xopJ5, xopAC, and xopR genes. The EMMA (Efficient Mixed-Model Association) model was used to run the association. Random effects kinship matrix was also used to controls for population structure. Corrections for multiple testing in order to remove false positives with FDR (false discovery rate) correction 10% were included.
Manhattan plot of EMMA describing the association between each trait and both markers. Numbers of significant markers in each trait are vary.
Decreasing number of sample are affected the significant AFLP detected in the associations. Less significant markers were detected when 37 strains for amylase and endoglucanase activities used. Different trend was shown for polygalacturonase, protease and swarming. Indeed, for these three traits and more importantly for protease activity, the number of significant markers obtained wit h AFLP 37 is higher than with AFLP 50. The result on swarming motility is not well based.
SNPs based phenotype tested seems give more precise relevancy than AFLP, but differences 1513 AFLP and 247480 SNPs were also contributed to the similar pattern of the Manhattan plot for swarming motility with SNPs markers as for AFLP markers were found about 12 significant SNPs markers for Amylase, 6 for endoglucanase, 50 for polygalacturonase, but non-significant for protease were detected. For both motilities, a high number of significant markers were detected i.e. 312 SNPs associated to swimming and 279 SNPs associated with swarming.
ORF (Open Reading Frame) sequence positions of the reference strain Xcc8004 to identify significant coding SNPs were used. Six significant markers obtained for the endoglucanase activity are coding for hypothetical proteins. Two of them are in the same operon. Two genes coding for hypothetical proteins were determined as significant, i.e. SNPs for amylase activity. Interestingly, four significant SNPs for amylase activity are clustered in the genome. These genes code for proteins with enzymatic activities. The role of these proteins in the secretion of extracellular enzymes like amylase and endoglucanase are presumably important. Among significant SNPs, 15 proteins, 8 of them being classified as signal peptide proteins, which mean they may be secreted by the Type II secretion system in Xcc.
Copyright© 2012, by Bogor Agricultural University All rights reserved
No part or all of this thesis may be excerpted without inclusion or mentioning the sources
a. Excerption only for research and education use, writing for scientific papers, reporting, critical writing or reviewing of a problem
GENOME-WIDE ASSOCIATION STUDY FOR NATURAL
VARIATION IN ENZYMATIC ACTIVITIES AND MOTILITY
TRAITS IN Xanthomonas campestris pv. campestris
NANI MARYANI
A thesis submitted as partial fulfillment of the requirement for the degree of Master of Science in Microbiology
GRADUATE SCHOOL
BOGOR AGRICULTURAL UNIVERSITY
BOGOR
Thesis External Examiner in INSA de Toulouse, France: Dr. Jean Michelle François
Dr. P. Soucaille Prof. Claude Maranges
Thesis Title : Genome-Wide Association Study for Natural Variation in Enzymatic Activities and Motility Traits in Xanthomonas campestris pv. campestris.
Name : Nani Maryani Student I.D. : G351107251
Approved Co-Major Professors,
Prof. Matthieu Arlat Dr. Ir. Lisdar I Sudirman
Dr. Ir. Gayuh Rahayu
Agreed
Coordinator of Microbiology Program Dean of Graduate School
Prof. Dr. Anja Meryandini, M.S. Dr.Ir. Dahrul Syah, M.Sc.Agr.
ACKNOWLEDGEMENT
In the name of Allâh the Most Gracious, The Most Merciful. Many individuals were responsible for the crystallization of this work, whose associations and encouragement have contributed to the accomplishment of the present report, and I would like to pay tribute to all of them.
I would like to share my deepest gratitude and sincere appreciation to my supervisors Prof. Matthieu Arlat, my Lab. supervisor Dr. Emmanuelle Lauber and Dr. Laurent Noël for their unwavering patience, advice, direct guidance and valuable support during my research at LIPM. I would also like to acknowledge Dr. Anne Geniselle, INRA Versailles, for her assistance and guidance on GWAS analysis in Paris. I must also thank the Directorate General of Higher Education (DIKTI), Ministry of National Education (Republic of Indonesia) for awarding of the Double Degree Indonesia-France (DDIP) Scholarship. I would also like to thanks to Dr. Ir. Lisdar I Sudirman and Dr. Ir. Gayuh Rahayu for their advice to finish the program at Bogor Agricultural University.
My sincere appreciation goes to all lab members Martine Lautier, Claudine Zischek, Fransky Hantelys, Thomas Dugé de Bernonville, Endrick Guy, and Brice Roux for assistance, willingness to share their knowledge, and friendship. I would also like to extend my appreciation to Prof. Claude Maranges of INSA Toulouse for his support and encouragement.
I would also like to thank Mohammad Belaffif for all the discussion time and PPI Toulouse for their support during my time in Toulouse. I would also like to thank Microbiology students IPB 2010 for their support and friendship during my time in IPB. Finally I would like to thank my parents and my sisters for their support and love was by far the most important element that allowed me to continue my studies.
Bogor, November 2012
AUTOBIOGRAPHY
The author was born on the 29th July 1983 from Mr. Martawi and Mrs. Endang Sunarti of Jakarta, Indonesia. She joined Bogor Agricultural University (IPB) and graduated with Bachelor Science in Biology in 2005. In 2006, she joined School of Universe (SOU) Parung Bogor as high school teacher on Biotechnology and Agriculture. She later joined University of Ibn Khaldun in 2007 where she completed program for License of Teaching in Biology Education.
She joined the Ministry of Education as a junior lecturer in the Department of Biology Education, University of Sultan Ageng Tirtayasa (UNTIRTA) Serang Banten in 2009. In August 2010, the author was awarded a scholarship for Master Degree, Double Degree Indonesia France (DDIP) Program by the Indonesian Government through the Directorate General of Higher Education (DIKTI), Ministry of Education. She joined Bogor Agricultural University in Microbiology Mayor Program.
TABLE OF CONTENTS
Page
LIST OF TABLES ... xiii
LIST OF FIGURES ... xiv
LIST OF ANNEXES ... xv
ABBREVIATIONS LIST ... xvi
INTRODUCTION ... 1
LITERATURE REVIEW Genome-Wide Association Studies (GWAS) ... 3
Xanthomonas campestris pv. campestris (Xcc) and its Virulence Factors ... 3
Genetic Markers ... 5
MATERALS AND METHODS Place and Duration of the Study ... 7
Bacterial Strains and Growth Conditions ... 7
Phenotypic Characters Assays ... 7
PCR (Polymerase Chain Reaction) ... 9
Statistical Analyses ... 10
RESULTS Variation on Enzymatic Activities and Motility of Xcc ... 13
Heritability of the Traits ... 14
Correlation among Traits ... 15
Connection between the Traits and the Phylogenetic Tree ... 15
GWAS ... 17
DISCUSSION ... 25
CONCLUSION AND PERSPECTIVE ... 29
LITERATURE CITED ... 31
LIST OF TABLES
Page
1 Primers of four Xcc effectors genes ... 9
2 Pearson's product-moment correlation between traits ... 16
3 Pearson's product-moment correlation between traits and clade... 16
4 Numbers of significant markers after FDR correction ... 20
LIST OF FIGURES
Page 1 Examples of degradation halos observed for different enzymatic
activities ... 13 2 Examples of motility halos. ... 14 3 Boxplot of phenotypic data on each trait. ... 15 4 Distribution of phenotypic results used for GWA studies in 50
Xcc strains. ... 18 5 Manhattan plots for enzymatic activities on 50 Xcc strains using
AFLP markers. ... 19 6 Manhattan plots for motility on 50 Xcc strains using AFLP
markers. ... 20 7 Manhattan plots for motility on 37 Xcc strains using SNPs
markers. ... 21 8 Manhattan plots for enzymatic activities on 37 Xcc strains using
LIST OF ANNEXES
Page
1 Name and Origins of the 50 Xcc Strains. ... 35
2 Manhattan Plot of GWAS on 37 Xcc strains using AFLP markers on enzymatic activities and motility ... 37
3 Heritability of each Traits on 50 Xcc Strains ... 38
4 Mean of Phenotypic Value on all Strains ... 42
ABBREVIATIONS LIST
ANOVA Analysis of Variance
AFLP Amplified Fragment Length Polymorphism CMC Carboxymethyl Cellulose
dNTP deoxyribonucleotide triphosphates
DNA Deoxyribonucleic acid
EMMA Efficient Mixed-Model Association FDR False Discovery Rate
GWAS Genome wide association study MAF Minor Allele Frequency
MB MegaBases
OD Optical Density
ORF Open Reading Frame
PCR Polymerase Chain Reactions PEG Polyethylene Glycol
QTL Quantitative Traits Loci
RFLP Restriction fragment length polymorphism
SNPs Single Nucleotide Polymorphisms T3E Type Three Effector
INTRODUCTION
A better understanding of the molecular genetics bases for phenotypic variation is one of the main challenges in modern biology (Aranzana et al. 2005). With the decreasing cost of genotyping, information of genotype data became more accessible and enables genome-wide approaches in most organisms. Genome-wide association studies (GWAS) has emerged as a promising tool to reveal the genetic basis of organisms.
For the last decade, most of the GWAS have been used to understand common human diseases. Researchers seek to identify regions of the genome where individuals that are phenotypically similar are also unusually closely related. In bref, a genome-wide association is a whole genome scan that tests the association between the genotypes at each locus and a given phenotype (Bergelson & Roux 2010).
Recently, GWAS has been used to identify common alleles of major effect of phenotypic variation in plants. Atwell et al. (2010) reported that studying 107 phenotypes of Arabidopsis thaliana using GWAS gave excellent candidate gene responsible for the traits variation. With the development and assembling technique from those of human GWAS, they also suggest that this approach can be feasible for many other organisms. These studies, owing to advances in genotyping and sequencing technology, become an obvious general approach for studying the genetic of natural variation of traits of agricultural importance (Atwell et al. 2010). However, to date, no GWAS has been reported for studying association between phenotype and genotype variations in bacteria.
on host plants. These characteristics led us to give more attention to study both phenotypic and genotypic variations in this pathogen. This first study emphasized two virulence traits of Xcc which are extracellular enzymes and motility.
Extracellular enzymes are one of the most important virulence factors in Xcc besides other factors such as avirulence genes (avr) and hrp genes. These enzymes are the first virulence factors that are secreted by the bacteria when they attack plants. The role of the extracellular enzymes in pathogenicity encouraged us to study both phenotype and genotype variation among strains of Xcc. Motility is another important pathogenicity factor in bacteria. Various forms of surface motility enable bacteria to establish symbiotic and pathogenic associations with plants (Rashid & Kornberg 2000).
LITERATURE REVIEW
Genome-Wide Association Studies (GWAS)
For the last two decades, GWAS has been used as a method to identify a multitude of subtle genetic effects increasing the risk of ‘complex’ disease and among unrelated individuals. Complex means that many genetic variants are contributing to the trait. GWAS has been used to study genetic variants associated with human disease such as obesity related traits (Scuteri et al. 2007), autoimmune disease (Pasaniuc et al. 2011), and also genetic determinants of complex diseases for the human population as a whole (Rosenberg et al. 2010). GWAS in humans use dense maps of SNPs that cover the genome, to look for allele-frequency differences between cases (patients with specific disease or individuals with certain trait) and controls. A significant frequency difference is considered to indicate that the corresponding region of the genome contains functional DNA-sequence variants that influence the disease or the trait in question (Kruglyak 2008). The goal of population association studies is to identify pattern of polymorphism that vary systematically between individuals with different disease states and could therefore represent the effects of risk-enhancing or protective alleles (Balding 2006).
GWAS are also used to study genetic variation in plants. These approaches successfully identified candidate genes on the model plant Arabidopsis thaliana (Aranzana et al. 2005, Atwell et al. 2010). Recently, Pasam et al. (2012) reported that GWA studies based on linkage disequilibrium provide a promising tool for the detection and fine mapping of complex agronomic traits. Genome-wide association mapping on plants are bringing a breath fresh air to the area of gene discovery. However, no GWAS are so far reported on microorganisms. It is interesting to perform GWAS to study phenotypic variation in Xcc.
Xanthomonas campestris pv. campestris (Xcc) and its Virulence Factors
blackened veins and V-shaped necrotic lesions at the foliar margin (Alvarez 2000). The ability of Xcc to elicit disease depends upon the synthesis of a number of factors, including, in addition to avirulence proteins (avr) and type III secretion system and effectors, the extracellular polysaccharide xanthan, extracellular plant cell wall degrading enzymes and cyclic glucan, as well as the formation of biofilms (Vojnov & Dow 2009).
Extracellular enzymes are one of the most important virulence factors in Xcc. Many extracellular enzymes, including pectin lyases, cellulases, and proteases are secreted during first attack to the plants (Aranzana 2000). The study of molecular determinants of pathogenicity of this bacterium has suggested a role for extracellular enzymes in the disease process. Dow et al. (1990) reported that mutants pleiotropically defective in the synthesis or export of cellulases, polygalacturonate lyases, proteases, and amylases are non-pathogenic in all plant tests used. Xcc produces protease, endoglucanase, polygalacturonate lyase, lipase, and amylase activities and all these enzymes have the capacity to degrade plant cell components (Dow et al. 1987). Therefore with the accessibility of genomic data on Xcc in the laboratory, this research started to study the natural genetic variation of 50 Xcc strains using GWAS approach for these phenotypes.
This research started to study GWAS with four extracellular enzymes produced by Xcc which are amylases, endoglucanases, polygalacturonases, and proteases. Protease is an essential factor at early stages of the disease process, but once infection is well advanced, the enzyme is less significant. Endoglucanase (“cellulase”) is the major extracellular protein produced by Xcc and has a structure typical of other prokaryotic endoglucanases (Gough et al. 1988). The role of this enzyme is not well understood but it may contribute to bacterial nutrition during the saprophytic phase of the life cycle. Polygalacturonase and amylase have not been studied in detail but both enzymes have an important role in the early steps of infection of the plant.
to translocate to preferred hosts and access to optimal colonization sites within them, and dispersal in the environment during the course of transmission. However, it is interesting also to include this phenotype on our study.
Genetic Markers
Genetic markers are distinctive features among individuals in the genetic map. In a geographical map, these markers are recognizable components of the landscape such as rivers, roads, and buildings. Instead of genes, many markers in a genetic landscape such as RFLPs (restriction fragment length polymorphisms), microsatelites, SNPs (single nucleotide polymorphisms) and AFLPs (amplified fragment length polymorphisms) can used.
A genetic marker provides informations about allelic variation at a given locus. These molecular markers have been applied to many biological questions, ranging from gene mapping to population genetics, phylogenetic reconstruction, paternity testing and forensic applications (Schötterer 2004). Genetic markers able to distinguish between genotypes that are relevant to a trait of interest are a key goal in genetics. Often, this distinction is not based directly on the trait of interest, but on informative marker systems (Schötterer 2004). Two different markers used in this study are AFLP and SNPs, as genotype data for the association studies.
AFLP. AFLPs are polymerase chain reaction (PCR)-based markers for the rapid screening of genetic variations. Because of their high replicability and ease of use, AFLP markers have emerged as a major new type of genetic markers with broad application in systematics, pathotyping, population genetics, DNA fingerprinting and quantitative trait loci (QTL) mapping. AFLP markers have proved useful for assessing genetic differences among individuals, populations and independently evolving lineages, such as species (Mueller & Wolfenbarger 1999). AFLP methods rapidly generate hundreds of highly replicable markers from DNA of any organism.
SNPs. SNPs or Single Nucleotides Polymorphisms are individual point mutations in the genome. These markers are commonly used as markers in GWA studies. Their abundant numbers make them good markers for the studies beside their lies in coding DNA (genes) and also non coding DNA. SNP detection is more rapid because it is based on oligonucleotide hybridization analysis. Efficient and cost-effective high-throughput SNP genotyping have been developed (Macdonald et al. 2005).
MATERIALS AND METHODS
Place and Duration of the Study
This research was carried out from November 2011 to May 2012 at Laboratoire Interaction Plant-Microorganism (LIPM-UMR CNRS INRA 2594/441) Toulouse, France. GWAS analyses was carried out at INRA (Institute Nationale de la Recheche Agronomique) Versailles, Paris France.
Bacterial Strains and Growth Conditions
50 strains of Xanthomonas campestris pv. campestris (Xcc) from LIPM collection were used in this study. Strains name and their origin are presented the Annexe 1. All the strains were maintained in glycerol 20% and reserved at -800C. Each week, strains were striked out from the stock to MOKA agar medium (Yeast Extract 4 g/L, Casamino acids 8 g/L, K2HPO4 2 g/L, MgSO4.7H2O 0.3 g/L, Bacto
Agar 15 g/L) (Blanvillain et al. 2007) and incubated at 280C for 3 days. These isolated strains were used as the source of inoculum in each experiment.
Phenotypic Characters Assays
Phenotypic characters tested in this study were enzymatic activities and motility.
Preparations of the bacterial solution for enzymatic tests. Two mL of overnight cultures of Xcc in MOKA liquid medium were harvested by centrifugation 4 min at 10 000 rpm. Pellets were resuspended in 1mL of MOKA broth. Concentration of bacteria was determined by measuring the optical density of the suspension with spectrophotometer 600nm. One ml of bacterial suspension at OD600 = 0.1 (108 CFU/mL) and 500 µ l of bacterial suspension at OD600 = 4
(4.109 cells/mL) were prepared in MOKA broth.
Enzymatic Activity Assays: radial diffusion assays method in agar plates containing substrate
Amylase Activity. 0.125% starch (Amilum potato starch, 2.5% w/v in H2O)
suspensions at OD600 4 were spotted on the plates that were incubated at 28°C for
24 hours. The plates were then stained with Lugol (iodine (I) 5g/L, potassium iodide (KI) 10g/L). Amylase activities of the colony were measure by the diameter of the Halo (clear zone) in the plates that present an intense blue color. Activity (arbitrary units) was calculated with the formula:
Endoglucanase Activity. 0.25% carboxymethyle cellulose (CMC) (w/v) in MOKA medium was used as substrate for endoglucanase activity. 5 L of bacterial suspension at OD600 4 were spotted on the plates and incubated at
28°Cfor 24 hours. The plates were then stained with Congo red 0.1% (w/v) for 1 hour and washed with NaCl 1M for 10 min. The clear zone around the colony indicates the degradation of CMC by endoglucanase. The activity was measured as described above.
Polygalacturonase Activity. 0.125% Polygalacturonic acid (Sodium polypectate 2,5% w/v in H2O, pH 7) in MOKA medium was used as substrate for
Polygalacturonase activity. 5 L of bacterial suspension at OD600 4 was spotted on
the plates that were incubated at 28°Cfor 24 hours. The plates were then washed with CTAB 1% (in water) for 30 minutes, stained with ruthenium red 0.1% (water) for 30 minutes and washed with water. Polygalacturonase activity was determined as described above.
Protease Activity. 5 L of bacterial suspension at OD600 4 were spotted on
MOKA plates containing 1% skimmed milk (w/v) and 100 µM FeSO4. The plates
were incubated at 28°C for 48 hours. Proteolytic activity was scored by detecting the degradation of the milk proteins seen as a zone of clearing around the colonies. The activity was calculated as described above.
Motility. 0.2% (w/v) agar in MOKA medium was prepared as medium for swimming and 0.7% agar for swarming. 2 L of bacterial suspensions at OD 0.1 were spotted in the center of the plates and incubated at 28°C. Diameter of
Arbitary unit of enzyme activities = ( φ Halo)2–(φ colon y)2
bacterial spreading in agar was measured as motile activity. Swimming activity was measured after 24h incubation and after 48h for swarming activity.
PCR (Polymerase Chain Reaction)
To verify that the strains were not inverted during the experiments, at the end of one replicate of protease activity, all the strains were verified by PCR using 4 couple of primers specific to xopAL2, xopJ5, xopAC, and xopR genes (Table 1). These genes code for type three effectors and their presence/absence in the 50 Xcc strains was already determined (E. Guy, PhD). PCR mix were prepared with 0.5 L dNTP 10 mM, 0.3 L forward primers 10 µM, 0.3 L reverse primers 10 µM, 0.2 L Taq polymerase 5 U/µL, 4 L green taq reaction buffer 5X, 10.8 L H2O and 4 L of bacterial suspensions in PEG (poly-ethylene-glycol)/KOH pH13.
PCR was set for 1 cycle at 95°C for 10 minutes (lysis cells and denaturation), 30 cycles 95°C (denaturation) for 30 sec, 48°C (hybridization) for 30 sec, 72°C (elongation) for 30 sec, 1 cycle at 72°C (post-PCR) for 10 min, and finally 1 cycle at 17°C (cooling down) for 10 min. PCR products were analyzed by separation on 1.5% agarose gelin TAE 1X (tris-acetic-EDTA, EDTA 1mM, tris 10mM, pH 8). DNA in agarose gels was stained with the fluorescent dye ethidium bromide and bands were visualized with UV light.
Table 1 Primers of four Xcc effectors genes
Statistical Analyses Phenotypic Data
Experimental Design. Each trait was measured using three technical replicates per experiment and three independent biological experiments. Randomization of samples was conducted both in technical and biological replicates. Strain Xcc8004 was always present in each experiment as a control strain and further used as a covariate in the model to calculate the fitted values of each trait.
Adjusted Means. First, the arithmetic mean was calculated among technical replicates of each strain and used this mean to calculate adjusted mean,
using the following ANOVA model with lm function in R: Yijk= + si+ ej+ j + ,
where Yijk is the trait (enzymatic activities or motilities) for strain i, experiment j; is the overall mean of trait; s is the strain effect, e is the experiment effect, is
covariate, and is the error. This model accounts for differences between the three
experiments. The adjusted means were used to run the association tests.
Genotypic Data. AFLP and SNP markers were used as genotype data obtained from bioinformatics information LIPM Toulouse, France. 50 Xcc strains were considered for the AFLP and 37 strains SNPs markers (Annexe 1). SNPs data were obtained from whole genome sequencing of 37 strains (Annexe 1) after alignment of each strain against the reference genome (Xcc8004). 1513 AFLP are considered to be polymorphic markers out of 1943 to run the GWAS, whereas 247480 polymorphic SNPs markers with a minor allele frequency ≥ 5% were used, for a total of 45953 regulatory SNPs and 201527 coding SNPs. To observe whether the sample size affects the number of significant tests, GWAS with 37 strain AFLP markers was also ran, using 1462 polymorphic markers.
http://darwin.cirad.fr/Home.php). It represents the values of similarity measures between all pairs of strains. The matrix was then used for association tests to correct for the relatedness among individuals.
GWAS. The EMMA (Efficient Mixed-Model Association) model was used to run GWAS (Kang et al. 2008). This model accounts for correction due to the structure of the sample. EMMA controls for population structure using matrix of random effects (kinship matrix). Linear function of EMMA:
Where Y, is the phenotype X is a vector of genotypes at marker being tested (AFLP or SNP markers), u and ε are random effects, which capture the variance due to background genetic factors and the environment. EMMA is implemented in an R software package (http:/mouse.cs.ucla.edu/emma
)
; for this association the function emma.ML.LRT was used.This analysis includes a lot of tests (there were as many tests as markers), thus it is necessary to include a correction for multiple testing in order to remove false positives (tests with p < 0.05 that are likely no true association, because the more tests performed the higher probability to meet false positive). To be cautious about false positive rate and also not neglect false negatives, FDR (False Discovery Rate) at 10% was used for the correction (Benjamini & Hochberg 1995).
RESULTS
Variation on Enzymatic Activities and Motility of Xcc
Enzymatic activities (Figure 1) and motility (Figure 2) has been successfully phenotyped, these traits are play an important role in pathogenicity of
Xanthomonas campestris (Ryan et al. 2011). Extracellular enzymes produced by
Xcc are able to degrade specific substrates added in plates. A collection of 50 strains of Xcc was tested and differences in the diameter of the halo (Figure 1) were measured for the different strains of Xcc and relative enzymatic activities were inferred from these observations. This suggests that each strain has different enzymatic activities. Similarly, swimming and swarming motilities were variable among the strains (Figure 2).
Figure 1 Examples of degradation halos observed for different enzymatic activities A. Amilase, B. Polygalacturonase, C. Protease, D. Endoglucanase.
A B
Figure 2 Examples of motility halos. A. swimming after 24h incubation (0.2% agar), B. swarming after 48h incubation (0.7% agar).
In all experiments, strain Xcc8004 was used as a covariate. Therefore, this strain was included in all plates to standardize the results. All of the strains were phenotyped with a total of nine replicates for each trait. Figure 3 shows boxplots result obtained for phenotypic data on each trait. Data dispersion (variation) was observed for all traits and some outliers were present such as in amylase, endoglucanase and swarming motility. These outlier data will be considered later in the manuscript to explain some of the results in association studies.
Heritability of the traits
This result also indicates that sufficient technical and biological replicates were performed.
A B
Figure 3 Boxplot of phenotypic data on each trait. A. Enzymatic activities, B. Motility (Y axis showed arbitrary unit of enzymes activities).
Correlation among Traits
Correlation patterns between traits based on Pearson’s correlation test shown in table 2. Some traits have positive correlation to the others. For instance, protease activity is positively correlated with polygalacturonase activity. These activities showed highly significant P-value (2.06e-07) besides the correlation value is high as well (0.6581045). It is interesting to see in the further study whether the markers associated with both enzymes are common. In contrary, protease activity is negatively correlated with endoglucanase activity. Amylase activity is also positively correlated with swimming and swimming is negatively correlated with swarming. However, for these three correlations, correlation values obtained are less significant.
Connection between the Traits and the Phylogenetic Tree
Population structure can generate spurious genotype-phenotype association. It is necessary to verify whether the phylogeny between the strains may give an effect on the traits. Therefore the corrections of relatedness among the strains are needed to avoid spurious associations. Table 3 showed Pearson’s correlation
between traits and clade. No significant correlation was found, except for swimming motility. Thus, one should be cautious interpreting GWAS results on this trait. Otherwise, mixed model approach that involves estimated kinship was used to limit/correct phylogeny effect in the association. Kinship matrix produced from dissimilarity matrix of AFLP markers was used in the association analysis.
Table 2 Pearson's product-moment correlation between traits
Amylase Endogluc Polygal Protease Swimming
Amylase Pearson 1
** Significance at 95% confidence interval.
Table 3 Pearson's product-moment correlation between traits and clade
Figure 4 shows the distribution of phenotypes against the phylogenetic tree of 50 Xcc strains. This phylogenetic tree was generated using 1513 AFLP neutral markers, and the genotype data generated using covariate models. Visually, the distribution of phenotypes does not seem to follow the phylogenetic tree except for swimming, which is in agreement with the results obtained by the Pearson’s correlation analysis (Table 3). These set of data were then used to run association studies.
GWAS
A genome-wide association study was performed using 1513 AFLP markers (when 50 strains were considered), 1462 AFLP markers (when 37 strains were considered), and 201527 SNPs markers. With AFLP markers, 50 strains could be included. For SNPs data extracted from whole genome sequencing projects, only 37 strains could be included. For AFLP markers, GWAS was performed with both datasets (50 and 37 strains). The same AFLP-derived Kinship matrix were included in the EMMA analysis for both marker sets.
Figure 4 Distribution of phenotypic results used for GWA studies in 50 Xcc strains. Colors degradation shows the level of activity from the lowest activity (white) to the highest activity (dark brown).
Figure 5 Manhattan plots for enzymatic activities on 50 Xcc strains using AFLP markers. Y axis showed the distribution of -log10 (P-value). X axis
showed the positions (random) of AFLP markers. The dashed red line shows FDR Threshold sig. P-value.
Manhattan plots describing associations for AFLP and motility traits are shown in Figure 6. Line pattern observed in EMMA results of swarming motility seems spurious. There could be several reasons to explain this line. Population structure effect might not been corrected enough and some outliers in the phenotypic data could also explain this result. EMMA results with 37 Xcc strains are shown in Annexe 2.
Figure 6 Manhattan plots for motility on 50 Xcc strains using AFLP markers. Y axis showed the distribution of -log10 (P-value). X axis showed the
positions (random) of AFLP markers. The dashed red line showed threshold for significant P-values after FDR correction.
Table 4 summarizes the number of significant markers for each trait passing FDR<10%. Data of both GWAS with AFLP showed that less significant markers are detected for 37 strains for amylase and endoglucanase activities. However, a different trend was shown for polygalacturonase, protease and swarming. Indeed, for these three traits and more importantly for protease activity, the number of significant markers obtained with AFLP 37 is higher than with AFLP 50. Further analyses are required to understand the reasons of these results.
It is interesting also to look of the suspicious result on swarming motility. The large number of significant markers (with P<10-5) for swarming is most likely due to an artefact: low variance was observed among most of the strains. Indeed only one strain had a high phenotypic value, thus many significant markers observed might be due to background genetic differences between this strain and the others (Annexe 3).
Table 4 Numbers of significant markers after FDR correction
Marker Amylase Endogluc Polygal Protease Swimming Swarming
AFLP 50 9 15 2 3 2 57
AFLP 37 3 8 54 231 3 86
SNP 12 6 50 - 312 279
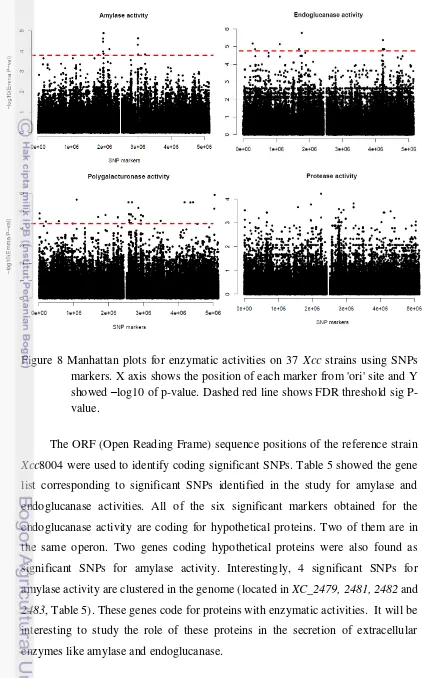
EMMA with SNP. The SNP panels used for EMMA only include the markers with MAF greater than 5%. Figures 7 and 8 show Manhattan plots of EMMA on motility and enzymatic activities traits, respectively. Effect of SNPs on phenotype tested seems to be higher than AFLP, but the huge differences in the number of markers that used on both analyses (1513 AFLP and 247480 SNPs) have to be considered. Obviously, similar pattern was found in the Manhattan plot for swarming motility with SNPs markers as for AFLP markers. It is not surprising since they came from the same set of phenotypic data.
Results obtained from SNPs after FDR corrections are showed in table 4. For enzymatic activities significant SNPs markers was found, 12 for Amylase, 6 for endoglucanase, 50 for polygalacturonase, but unfortunately nothing significant for protease. For both motilities a high number of significant markers were obtained, 312 SNPs associated to swimming and 279 SNPs associated with swarming. For swarming, other factor have to be considered like genetic differences of the outlier strain, and not true associations.
Figure 7 Manhattan plots for motility on 37 Xcc strains using SNPs markers. X axis shows the position of each marker from 'ori' site and Y showed – log10 of p-value. Dashed red line shows FDR threshold sig P-value.
Figure 8 Manhattan plots for enzymatic activities on 37 Xcc strains using SNPs markers. X axis shows the position of each marker from 'ori' site and Y showed –log10 of p-value. Dashed red line shows FDR threshold sig P-value.
Table 5 Characteristics of significant coding SNPs on endoglucanase and amylase activities
Positions P-value Trait Sequence Product Name PsortII
285213 6.8e-06 endogluc XC_0236
hypothetical protein XC_0236a
Signal peptide
1104278 7.3e-06 endogluc XC_0920
hypothetical protein
1768256 1.6e-06 endogluc XC_1466
hypothetical protein XC_1466a
clp protease/periplasmicb
Signal peptide
1768271 1.6e-06 endogluc XC_1466
hypothetical protein XC_1466a
Signal peptide
4223898 4.4e-06 endogluc XC_3558
hypothetical protein XC_3558a Protein
kinaseb -
4264570 1.4e-05 endogluc XC_3587
hypothetical protein XC_3587a Putative ATPase/ specific X.
campb -
1954981 3.4e-05 amylase XC_1628
excinuclease ABC
subunit Ba -
1964734 1.2e-05 amylase XC_1639
VirB4 proteina
Export ATPaseb -
1965038 1.2e-05 amylase XC_1639 VirB4 proteina - 1965356 1.2e-05 amylase XC_1639 VirB4 proteina -
2143006 8.1e-05 amylase XC_1762
formate dehydrogenase a
3004423 2.3e-05 amylase XC_2481
hypothetical protein
3006909 2.3e-05 amylase XC_2482
sialic acid-specific
9-O-3008805 2.3e-05 amylase XC_2483
Cellulasea a Xcc8004 genome annotation
DISCUSSION
This study shows that GWA study is feasible also to study variations in bacterial pathogens like Xcc using 6 traits for a sample of 50 Xcc strains. This study obtained interesting results on genome-wide association using six traits in Xcc. This approach is commonly used to study human diseases and variation of complex traits in plant, and appears to be very useful to understand the genetic basis of pathogenicity of bacterial pathogens.
First results showed that there is substantial phenotypic variation among the 50 Xcc strains, showed by differences in activity on extracellular enzymes and motility. These strains are spreading in the different clade in phylogenic tree, and exhibit differences also in host specificity. Differences between Xanthomonas spp. in the repertoire of the genes that encode enzymes involved in the degradation of the plant cell wall might reflect differences in cell wall composition among the respective host or tissues and could also be related to differences in the symptoms produced by infection (Rashid & Kornberg 2000).
Some significant SNPs markers (with an FDR correction at 10%) were also detected for each trait except for protease activity. SNPs are common markers used for association studies from human to plants, and this study showed that they can be used in bacteria as well. With the huge number of SNPs markers used in this study, cautiousness for some significant associations in the result has to be considered. One fundamental problem is that the genome is so large that the pattern that are suggestive of a causal polymorphism could well arise by chance. To distinguish causal from spurious signals, tight standards for statistical significance need to be established (Balding 2006). Any given variant (or set of variants) is highly unlikely, a priori, to be causally associated with any given phenotype under the assumed model. Therefore strong evidence is required to overcome the appropriate skepticism about an association, one of the approach is to monitor the false discovery rate (FDR) which is the proportion of false positive test results among all positives (Benjamini & Hochberg 2005). FDR was chosen for correction instead of other methods because with the large scale SNP-association studies, multiple tests are a bane of statistical result.
potential GWAS in A. thaliana, and other species with similar pattern of variation (Aranzana et al. 2005).
CONCLUSION
Genome-wide association study was successfully performed on Xanthomonas campestris pv. campestris. A population of 50 Xcc strains was phenotyped on their enzymatic activities and motility. Using two different data sets of markers, some significant markers associated with the given phenotype were observed. Correction of population structure was also taken into account. AFLP markers were detected in all traits. Significant markers for amylase and endoglucanase activities using AFLP markers were decreased when the association was performed using 37 Xcc strains instead of 50 strains. With SNPs markers, hundreds significant markers were detected for motility and no significant SNPs marker was detected for protease activity. Identification of significant coding SNPs for amylase and endoglucanase using sequence positions of the reference strain Xcc8004 showed that some hypothetical proteins are associated with these enzymatic activities.
PERSPECTIVE
LITERATURE CITED
Alvarez A. 2000. Black rot of crucifers, pp. 21-52. In A.J. Slusarenko (ed.), Mechanisms of resistance to plant diseases. Kluwer Academic Publications, Dordrecht, Netherlands.
Aranzana MJ et al. 2005. Genome-Wide Association Mapping in Arabidobsis Identifies Previously Known Flowering Time and Pathogen Resistance Genes. PLoS Genet 1:0531-0538.
Atwell S et al. 2010. Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature 465:627-631.
Balding DJ. 2006. A tutorial on statistical methods for population association studies. Nat Rev Genet 7:781-791.
Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society 57:289-300.
Bergelson J, Roux F. 2010. Towards identifying genes underlying ecologically relevant traits in Arabidopsis thaliana.Nat Rev Genet 11:867-879.
Blanvillain S et al. 2007. Plant carbohydrate scavenging through tonB-dependent receptors: a feature shared by phytopathogenic and aquatic bacteria. PloS one 2:e224.
Dow JM, Scofield G, Trafford K, Turner PC, Daniels MJ. 1987. A gene cluster in Xanthomonas campestris pv. campestris required for pathogenicity controls the excretion of polygalacturonate lyase and other enzymes. Physiological and Molecular Plant Pathology 31:261-271.
Falconer DS, Mackay TFC. 1996. Introduction to quantitative genetics, 4th ed. Longman Scientific & Technical, Burnt Mill, Harlow, United Kingdom.
Gough C, Dow L, Maxwell J, Barber CE, Daniels MJ. 1988. Cloning of two Endoglucanase gene Xanthomonas campestris pv. campestris. Molecular Plant-Microbe Interactions 275-281.
Kang HM et al. 2008. Efficient control of population structure in model organism association mapping. Genetics 178:1709-1723.
Macdonald SJ, Pastinen T, Genissel A, Cornforth TW, Long AD. 2005. Low cost production-level SNP genotyping for large-scale association mapping applications. Genome Biology 6.
Mueller U, Wolfenbarger L. 1999. AFLP genotyping and fingerprinting. Trends in ecology & evolution 14:389-394.
Olsen KM et al. 2004. Linkage disequilibrium mapping of Arabidopsis CRY2 flowering time alleles. Genetics 167:1361-1369.
Pasaniuc B et al. 2011. Enhanced statistical tests for GWAS in admixed populations: assessment using African Americans from CARe and a Breast Cancer Consortium. PLoS genetics 7:e1001371.
Rashid MH, Kornberg A. 2000. Inorganic polyphosphate is needed for swimming, swarming, and twitching motilities of Pseudomonas aeruginosa. Proceedings of the National Academy of Sciences of the United States of America 97:4885-4890.
Rosenberg NA et al. 2010. Genome-wide association studies in diverse populations. Nat Rev Genet 11:356-366.
Ryan RP et al. 2011. Pathogenomics of Xanthomonas: understanding bacterium– plant interactions. Nat Rev Micro 9:344-355.
Schlötterer C. 2004. The evolution of molecular markers--just a matter of fashion? Nature reviews. Genetics 5:63-69.
Scuteri A et al. 2007. Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS genetics 3:1200-1210.
Code
N4 HRI3880 1975 Australia B oleracea capitata Cabbage
N5 HRI7758 1999 Brazil B oleracea tronchuda NA
N9 CFBP1119 1967 NA B oleracea var botrytis Cauliflower
N10 HRI1279A 1984 UK B oleracea capitata Cabbage
N11 HRI3883 1966 USA Raphanus sativus Radish
N12 CFBP1712 1975 France B oleracea capitata Cabbage
N13 CN17 2003 China B rapa subsppekinensis
Chinese cabbage
N14 CFBP4953 1999 Belgium B oleracea var botrytis Cauliflower
N15 CN11 2002 China B oleracea capitata Cabbage
N16 CN19 NA China NA NA
N17 HRI6189 1996 Portugal B oleracea var italica Broccoli
N18 CFBP1124 1967 France B oleracea var botrytis Cauliflower
N19 CN05 2002 China B rapa subsppekinensis
Chinese cabbage
N20 CFBP6865R 1975 Australia B oleracea capitata Cabbage
N21 CFBP5683 1979 France B oleracea NA
N24 CN03 2002 China B rapa subsppekinensis
Chinese cabbage
N25 CN15 2003 China B rapa subsppekinensis
Chinese cabbage
N26 HRI3811 NA USA B oleracea NA
N27 CN01 2001 China B rapa subsppekinensis
Chinese cabbage
N28 CFBP5130 2000 NA B oleracea NA
N29 CN10 2002 China B rapa subsppekinensis
Chinese cabbage
N30 CN20 2003 China B oleracea alboglabra Chinese kale
NA= Not Analyze.
N31 CN07 2002 China B rapa subsppekinensis
Chinese cabbage
N32 CN13 NA China NA NA
N33 CFBP4954 1999 Belgium B oleracea var botrytis Cauliflower
N34 CFBP1713 1975 France B oleracea var botrytis Cauliflower
N35 CN18 2003 China B juncea var foliosa Leaf mustard
N36 HRI6185 1996 Portugal B rapa
Turnip mustard
N37 CN16 2003 China B rapa subsppekinensis
Chinese cabbage
N38 HRI7283 1997 UK Cruciferous weed NA
N39 CFBP1869 1976 Ivory coast B oleracea NA
N40 HRI7805 1998 South Africa B oleracea var botrytis Cauliflower
N41 Xcc147 NA Brazil NA NA
N42 CFBP5817 2001 Chilli B oleracea var botrytis Cauliflower
N43 CFBP4956 1999 Belgium B oleracea var botrytis Cauliflower
N44 HRI3851A 1989 Greece B oleracea var botrytis Cauliflower
N45 CFBP6863 1958 Germany B oleracea capitata Cabbage
N46 HRI6382 1953 Canada B rapa
Turnip mustard
N47 CFBP4955 1999 Belgium B oleracea var botrytis Cauliflower
N48 HRI6412 1997 France B oleracea var botrytis Cauliflower
N49 CN06 2002 China B rapa subsppekinensis
Chinese cabbage
N50 CN02 2002 China B rapa subsppekinensis
Annexe 2 Manhattan Plot of GWAS on 37 Xcc strains using AFLP markers on enzymatic activities and motility
Distribution of -log10 (P-value) on Enzymatic activities (37 Xcc strains) (Y axis).
X axis showed the positions (random) of AFLP markers. The dashed red line showed threshold for significant P-values after FDR correction.
N30 7,485 0,876 8,483 0,992 8,383 0,981 N31 7,504 0,878 8,463 0,990 7,887 0,923 N32 -12,081 -1,413 5,663 0,662 8,044 0,941 N33 6,136 0,718 8,114 0,949 8,493 0,993 N34 8,397 0,982 8,432 0,986 8,207 0,960 N35 7,847 0,918 8,478 0,992 7,989 0,935 N36 6,335 0,741 8,542 0,999 8,548 1,000 N37 7,973 0,933 8,457 0,989 8,368 0,979 N38 8,532 0,998 5,135 0,601 8,544 0,999 N39 6,088 0,712 6,321 0,739 8,485 0,993 N40 2,682 0,314 2,448 0,286 8,543 0,999 N41 8,548 1,000 -5,138 -0,601 7,962 0,931 N42 8,036 0,940 8,517 0,996 8,545 1,000 N43 8,420 0,985 5,651 0,661 8,512 0,996 N44 7,873 0,921 8,496 0,994 8,549 1,000 N45 6,465 0,756 3,484 0,407 8,539 0,999 N46 7,792 0,912 6,379 0,746 7,776 0,910 N47 3,292 0,385 6,457 0,755 8,157 0,954 N48 6,719 0,786 8,249 0,965 8,464 0,990 N49 8,446 0,988 3,437 0,402 8,548 1,000 N50 8,499 0,994 1,802 0,211 8,493 0,993
Average 0,567 0,824 0,974
N30 8,537 0,999 8,549 1,000 8,538 0,999 N31 8,359 0,978 8,532 0,998 8,548 1,000 N32 8,548 1,000 8,532 0,998 8,548 1,000 N33 8,528 0,998 8,472 0,991 8,549 1,000 N34 8,463 0,990 8,489 0,993 8,548 1,000 N35 8,412 0,984 8,523 0,997 8,543 0,999 N36 8,512 0,996 8,191 0,958 8,542 0,999 N37 5,401 0,632 8,543 0,999 8,548 1,000 N38 8,547 1,000 8,549 1,000 8,547 1,000 N39 8,534 0,998 8,543 0,999 8,549 1,000 N40 8,506 0,995 8,545 1,000 8,549 1,000 N41 8,514 0,996 8,490 0,993 8,545 1,000 N42 8,549 1,000 8,538 0,999 8,548 1,000 N43 8,494 0,994 8,548 1,000 8,548 1,000 N44 8,549 1,000 8,542 0,999 8,548 1,000 N45 8,187 0,958 8,549 1,000 8,547 1,000 N46 8,549 1,000 8,548 1,000 8,548 1,000 N47 8,549 1,000 8,547 1,000 8,545 1,000 N48 8,541 0,999 8,475 0,991 8,547 1,000 N49 8,539 0,999 8,549 1,000 8,548 1,000 N50 8,112 0,949 8,546 1,000 8,548 1,000
Average 0,975 0,996 1,000
Annexe 4 Mean of Phenotypic Value on all Strains
N30 8,284 8,034 3,001 1,935 0,770 0,590
N31 8,255 7,402 3,927 2,375 1,365 0,581
N32 9,655 5,954 3,083 3,460 0,661 0,555
N33 9,247 8,346 1,075 0,315 1,883 0,571
N34 8,580 7,430 1,777 0,306 1,823 0,529
N35 6,926 8,187 4,359 1,131 1,211 0,636
N36 11,584 8,881 2,990 1,012 1,699 0,594
N37 10,014 6,197 3,944 3,034 1,705 0,554
N38 7,986 7,910 2,944 0,931 0,792 0,544
N39 8,077 7,663 2,484 0,250 0,567 0,612
N40 9,016 8,296 2,807 0,849 1,102 0,531
N41 9,628 8,168 2,497 2,676 0,733 0,613
N42 7,138 6,814 1,446 -0,070 0,923 0,598
N43 8,547 7,352 3,000 2,631 1,241 0,617
N44 7,752 6,377 0,233 0,109 1,094 0,559
N45 8,231 7,447 0,893 0,919 0,726 0,573
N46 9,539 6,509 3,980 3,086 1,810 0,548
N47 8,887 7,645 0,436 0,047 1,250 0,655
N48 8,158 6,926 1,457 0,170 1,438 0,544
N49 6,667 6,133 1,086 0,980 0,564 0,618
Annexe 5 Profile of the Laboratory
Laboratoire des Interaction Plantes Micro-organismes INRA (Institute Nationale des Recherche Agronomique) Toulouse. The Laboratory of Plant-Microbe Interactions (LIPM), created in 1981, is a combined INRA-CNRS Research Unit, attached to the CNRS Institutes of Biological Sciences (INSB) and Ecology & Environment (INEE), as well as to the INRA Departments of Plant Health & Environment (SPE) and Genetics & Plant Breeding (GAP). The LIPM is also part of a Research Federation (FR 3450, ex IFR 40, Agrobiosciences, Interactions and Biodiversity which is located on the INRA campus at Auzeville-Tolosan, south east of Toulouse.
LIPM has focused its research on the interactions between plants and either symbiotic or pathogenic microbes, coordinating studies on both plant and microbial partners. This research is performed using a small number of model species (Arabidopsis thaliana, Medicago truncatula). The studies are to address key biological questions concerning the determinants which control pathogenic/symbiotic associations, the mechanisms of host infection, inter- and intra-organism signaling, developmental programs, regulation of gene expression and finally the mechanisms of metabolic adaptation. Bacterial and plant genome organization is also a theme that are actively studied using both bioinformatics and genomic approaches.
To achieve the goals, LIPM has divided into two major research topics which concern root endosymbioses and plant-pathogen environment interactions. The LIPM addresses scientific questions which have a major impact on agriculture and the environment. In the long-term, the research aims to provide new concepts and tools in the struggle against bio-aggressors. Symbiotic interactions play an essential role in the provision of essential nutrients to legume plants, the importance of which is now being rediscovered in agronomical, economic, environmental and nutritional contexts.
regulation of symbiotic infection and nodule development, 4) Ralstonia pathogenicity determinants and their plant targets, 6) Infectious strategies of Xanthomonas, Resistance, 7) Susceptibility and cell death in Arabidopsis thaliana in response to bacterial pathogens, and 8) Genetics and genomics of abiotic and biotic stress responses.
Team “Infectious Strategies of Xanthomonas”. Team Infectious Strategies of Xantomonas is leading by Matthieu Arlat and Emmanuelle Lauber. In the team of Infectious strategies of Xanthomonas, the aim is to unravel the molecular mechanisms allowing the adaptation of phytopathogenic bacteria to their hosts with the long-term objective to propose innovative strategies to contend plant diseases.
The work is centered on the plant pathogenic bacterium Xanthomonas campestris pv. campestris (Xcc) which is the causal agent of black rot of crucifers. This bacterium infects plant of agronomical interest such as cabbage, broccoli or cauliflower as well as the model plant Arabidopis thaliana. Xcc belongs to a large genus which comprises 27 species that collectively affect more than 400 plant hosts. The genome sequences of 16 Xanthomonas strains affecting economically important crops, such as, rice, citrus, banana, tomato, pepper and bean are publicly available making this genus a good model for comparative genomics.
Two major determinants controlling the interaction with plants which are type III secretion effectors (T3Es) and Carbohydrate utilization systems (CUT) are focused on the studies. Studies on the genomic diversity of Xcc using phenotypic approaches are also starting recently. Indeed, 50 strains of Xcc are available and 40 strains already sequenced. Several approaches ranging from molecular biology, genetics, biochemistry and genomics are combining together to carry out the goal of the research.
http://www6.toulouse.inra.fr/lipm_eng/Research/Emmanuelle-Lauber-and