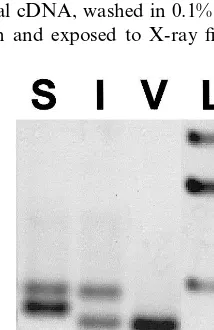Molecular characterization of the anthocyanidin synthase gene in
Forsythia
×
intermedia
reveals organ-specific expression during
flower development
Carlo Rosati
a,1, Alain Cadic
a, Michel Duron
a,*, Mathieu Ingouff
a,2,
Philippe Simoneau
baINRA C.R.Angers,Unite´ d’Ame´lioration des Espe`ces Fruitie`res et Ornementales,42Rue G.Morel,49071Beaucouze´ Cedex,France bLaboratoire Interactions Plantes-Microorganismes,Uni6ersite´ des Sciences d’Angers,2Boule6ard La6oisier,49045Angers,France
Received 3 March 1999; received in revised form 19 July 1999; accepted 20 July 1999
Abstract
The study of the anthocyanidin synthase (ANS) gene, one of the late structural genes in the anthocyanin pathway, was undertaken inForsythia×intermediacv. ‘Spring Glory’. Previous molecular and biochemical studies had demonstrated expression and activity of genes and enzymes upstream of ans. The ans gene was cloned and shown to be present in two copies in the
Forsythia genome. Expression analyses carried out on flower organs showed thatanswas expressed exclusively in anthocyanin-containing sepals and not in anthocyaninless anthers and petals.ansexpression in sepals showed induction of transcription at early flower developmental stages. Inspection of theanspromoter region as far as 790 bp upstream of the start codon revealed several potential DNA – protein binding motifs. The results from this paper, combined with previous data, show that the lack of
ansexpression should be the major cause of the absence of anthocyanins inForsythiapetals, thus providing directions for genetic engineering of flower color in this ornamental species. © 1999 Elsevier Science Ireland Ltd. All rights reserved.
Keywords:Anthocyanins; Flavonoid pathway; Flowers;Forsythia; Gene expression; Promoter
www.elsevier.com/locate/plantsci
1. Introduction
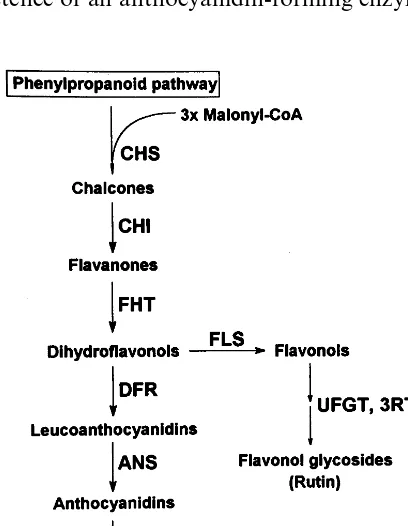
Anthocyanins are flavonoid pigments commonly found in plants, where they contribute to flower pigmentation and other functions as insect and animal attractants for flower pollination or fruit and seed dispersal. The structural genes of the flavonoid pathway (Fig. 1) have been cloned and characterized thoroughly in a number of plant
species [1]. Several regulatory factors, exerting their role spatially and developmentally on differ-ent sets of flavonoid pathway genes, have been identified also and studied ([2 – 4] and Refs. therein).
Forsythia (Oleaceae) is a popular yellow-flow-ered ornamental shrub, whose petals and anthers accumulate carotenoids, and whose vegetative or-gans (leaves, stems) and sepals accumulate an-thocyanins. We studied the flavonoid pathway in
Forsythia, to determine the causes for the lack of anthocyanins in petals and to eventually
comple-ment it by genetic engineering. Forsythia petals
contain huge quantities of flavonol glycosides, mainly rutin (quercetin 3-rutinoside) [5], which indicates that anthocyanin pathway enzymes as far as flavanone 3-hydroxylase (FHT; EC 1.14.11.9), and both glucosyl- (UFGT; EC 2.4.1.91) and
The reported nucleotide sequence data have appeared in the EMBL Nucleotide Sequence Database under the accession numbers Y12489 (ansmRNA) and Y13435 (anspromoter).
* Corresponding author. Tel.: +33-2-41225774; fax: + 33-2-41225755.
E-mail address:[email protected] (M. Duron)
1Present address: ENEA Casaccia – INN-BIOAG-BIMO, S.P.
026-301, via Anguillarese, 00060 S. Maria di Galeria, Rome, Italy.
2Present address: Department of Forest Genetics, Swedish
Univer-sity of Agricultural Sciences, P.O. Box 7027, S-75007 Uppsala, Swe-den.
rhamnosyl- (3RT) transferases, are active (Fig. 1). We demonstrated chalcone synthase (CHS; EC 2.3.1.74), FHT and flavonol synthase (FLS)
activ-ity in petals of Forsythia×intermedia cv. ‘Spring
Glory’ [5]. We also showed that expression of the dihydroflavonol 4-reductase (DFR; EC 1.1.1.219) gene is blocked in anthers but not in petals [6].
Moreover, petals of transgenic CaMV 35S::dfr
plants had increased dramaticallydfr mRNA
con-centration and DFR activity, but displayed a wild-type phenowild-type [6]. Therefore, we had concluded
that dfr is not responsible for the absence of
anthocyanins inForsythiapetals. Since activities of
all anthocyanin pathway enzymes as far as DFR were demonstrated, and glycosyltransferase activ-ity was implied by the major presence of
glycosy-lated flavonoids, the anthocyanidin synthase
(ANS) gene was likely to be expressed
differen-tially inForsythia flower whorls, and in particular
not expressed in petals.
ans, the last structural gene in the anthocyanin
pathway before glycosylation [1,2], encodes a protein belonging to the class of 2-oxoglutarate dependent dioxygenases (2-ODDs). ANS shares consistent homology with two other 2-ODDs of the flavonoid pathway, FHT and FLS [1,2]. The existence of an anthocyanidin-forming enzyme
act-ing between DFR and glycosyltransferases was supposed long before the molecular characteriza-tion of its genes [7,8]. Nevertheless, ANS has been
characterized only recently [1], and ans sequences
have become increasingly available [9 – 12].
In this work, the ans gene was cloned and its
expression studied in F.×intermedia cv. ‘Spring
Glory’ during flower development. The genomic
organization of the ansgene was investigated, and
the reported hybrid origin of F.×intermedia was
further supported by comparing ans sequences of
F.×intermedia with those of putative parental
species. Finally, the promoter region of the ans
gene was also cloned and its sequence scanned for the presence of DNA – protein binding motifs con-tained in a transcription factor database [13].
2. Materials and methods
2.1. Plant material
F.×intermedia cv. ‘Spring Glory’ plants were field grown in the INRA Angers collection. An-thers, petals and sepals were harvested from flower buds at different developmental stages (0, full winter dormancy, 45 days before start of anthesis; 1, 15 days before start of anthesis; 2, start of anthesis, petals 1 mm out of flower buds; 3, 3 days after start of anthesis, petals 2 – 3 mm out of flower buds; 4, 7 days after start of anthesis, petals
]5 mm out of flower buds) as described
previ-ously [6]. Young leaves of F.×intermedia cv.
‘Spring Glory’, F. suspensa var. fortunei and F.
6iridissima were collected from either greenhouse plants or in vitro cultures. Samples were stored at
−80°C until use.
2.2. DNA and RNA extraction and analysis
Total DNA was extracted from leaves as de-scribed previously [14]. Total RNA was extracted from flower organs and checked for purity and integrity as reported [6]. Southern and Northern
analyses were performed using 10-mg nucleic acid
samples according to standard techniques [15].
2.3. Cloning and analysis of ans coding sequence and promoter
All PCR primers were purchased from
GENOSYS (Pampisford, UK). Degenerate
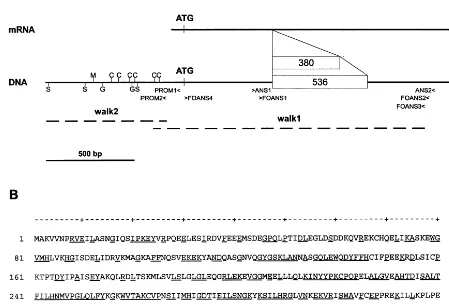
Fig. 2. (A) Schematic representation of the anslocus with the two alleles of F.×intermedia cv. ‘Spring Glory’. White boxes: introns (length shown in bp). Positions of initiation and stop codons, PCR primers (see text) and putative binding sites for transcription factors (C, C1/P; G, G box; M, MYB.Ph3; S, SBF-1) are indicated. Mapping of genomic walking fragments (walk 1 and walk 2, dashed lines) is indicated also. (B) Amino acid sequence deduced fromanscDNA ofF.×intermedia cv. ‘Spring Glory’. Underlined letters: consensus sequence for ten ANS sequences of dicotyledons.
primers (ANS1 5%
-AGC-AAG-TTH-GCM-AAY-ART-GC and ANS2 5%
-CCB-CTA-TGR-AGD-ATR-CTB-TT, Fig. 2), designed from multiple sequence alignments, were used to amplify the
correspondingForsythia ans cDNA fragment from
sepal first strand cDNA. Specific primers
(FOANS2 5%
-TCC-ATT-GGA-CAA-AAT-CTC-AAT-AG and FOANS3 5%
-CTC-AAT-AGT-ATC-TCC-AAT-GTG-C; PROM1 5%
-GGT-TCT-TGT-CCT-TGC-CTA-ATC-TTC-GGC-TG and
PRO-M2 5%
-CAA-CTT-TGT-CCC-ATT-ATT-CAT-AGC; Fig. 2) were designed to walk upstream as
far as the promoter region of ans, using a
PCR-based approach adapted from Ref. [16]. Briefly,
genomic DNA (2 mg) was digested overnight with
Ecl136II, EcoRV, SmaI, SspI or StuI restriction
enzymes. A fivefold molar excess of Adaptor [16] was ligated to each digested DNA sample. Two successive 30-cycle PCRs were carried out on these
samples using Adaptor-specific AP1 and AP2 for-ward primers [16] and either FOANS2 and
FOANS3 or PROM1 and PROM2 reverse
primers. Sequence homology analyses were carried
out using the BESTFIT program of the GCG
soft-ware package. The ans promoter sequence was
scanned for DNA – protein binding sites with
MATINSPECTOR program [13] using standard
set-tings (core similarity, 0.80; matrix similarity, 0.85).
2.4. Re6erse transcription (RT)-PCR
Total RNA (1 mg) was reverse transcribed using
anchored oligo-d(T) [17] primer and RT-PCRs
were carried out as reported [6]. FOANS1 (5%
-TGA-GTG-GCA-GGA-CTA-TTT-CTT-CC) and FOANS2 (Fig. 2), and FOEF1 and FOEF2 [6]
were used to amplify ans and elongation factor-1
Mini-Cycler™ (MJ Research) thermocycler program
was: initial denaturation 94°C/5 min; 30 cycles of
94°C/30 s, 55°C/30 s, 72°C/1 min; final extension
72°C/15 min.
3. Results
3.1. Analysis of ans coding sequence
A 491-bp RT-PCR product was amplified from sepal first strand cDNA using ANS1 and ANS2 degenerate primers and cloned. Based on its se-quence, FOANS2 and FOANS3 primers were
de-signed and used to amplify the 5% region of the
gene from small-scale genomic libraries. A ca. 1.6-kb fragment (walk 1, Fig. 2) was thus
am-plified from SmaI library. This fragment
over-lapped the previous ANS1 – ANS2 product and
spanned the 5% region of theansgene as far as the
−174 bp position in the promoter region (Fig. 2).
This 1.6-kb genomic clone contained a 536 bp intron (Fig. 2), located in the same position as that of Petunia [10] and Arabidopsis (GenBank
AL031018) ans. Alignment of this 1.6 kb clone
with other anscoding sequences revealed an ORF
having its ATG start codon in the same position asVitis[11]. Based on such data, FOANS4 primer
(5%
-ATG-GCT-AAG-GTG-GTG-AAT-CCG-AGA-G, Fig. 2) was used to amplify theForsythia
ans coding sequence and untranslated 3% region
from sepal first strand cDNA in a single-round
3%RACE experiment [18]. The resulting Forsythia
ans mRNA (GenBank Y12489) contains an ORF
of 1062 nucleotides encoding a polypeptide of 354 aminoacids (Fig. 2). Compared with all available
ans coding sequences, nucleotide sequence identity
varied between 60.0% (O. sati6a) and 75.5% (P.
hybrida); amino acid sequence similarity ranged
within 70.6% (O. sati6a) and 88.6% (P. hybrida)
with 47.5 and 77.6% of identity, respectively.
3.2. Genomic organization of ans
Southern analysis was carried out to assess the
number of ans copies, using restriction enzymes
expected not to cut in the genomic ans sequence.
Autoradiography displayed a two-band pattern for each restriction digestion (Fig. 3). The same
experiment carried out with an ans promoter (−
790 to −117 position) probe confirmed previous
results (data not shown). To determine whether
the two bands represented different ans genes or
alleles, PCRs were carried out on genomic DNA using FOANS1 and FOANS2 primers, flanking a region previously reported to contain an intron.
Two ans fragments of 969 and 813 bp were
am-plified (Fig. 4), differing only in the size of their introns (536 and 380 bp long, respectively), as analysis of their sequence revealed. Therefore, the bands of the Southern analysis should represent
two ans alleles. The same PCR experiment was
also carried out on reported parental genotypes of
F.×intermedia, F. 6iridissimaand F. suspensa var.
fortunei (Fig. 4). One single 813-bp PCR product,
Fig. 3. Southern analysis ofansinForsythia×intermediacv. ‘Spring Glory’. Genomic DNA was digested by EcoRI (E),
HindIII (H) andXbaI (X). Filter was hybridized overnight at 65°C with labeled FOANS1 – FOANS2 RT-PCR product am-plified from sepal cDNA, washed in 0.1% SSPE, 0.1% SDS at 65°C for 15 min and exposed to X-ray film for 4 days.
Fig. 4. Polymorphism ofans alleles of F.suspensa var. for-tunei (S), Forsythia×intermedia (I) and F. 6iridissima (V),
Fig. 5.ansexpression analysis in flower organs ofForsythia×
intermedia cv. ‘Spring Glory’ by RT-PCR. A, anthers; P, petals; S, sepals. Figures indicate flower developmental stages (see Section 2) [6]. Theef-1a gene was used as the
constitu-tively expressed control [6,18].
bp band of the constitutively expressedef-1agene,
used as positive internal control [5,18] (Fig. 5).
Northern analysis ofans expression in sepals (Fig.
6) showed thatans expression peaks at early stage
1 (2 weeks before the appearance of petals out of
flower buds) and decreases during flower
development.
Further sequencing of the ans gene allowed the
cloning of a ca. 700-bp DNA fragment in the promoter region (walk 2, Fig. 2). Its sequence overlapped that of the previous 1.6-kbp clone over
57 bp, and spanned the −790 to −123 positions
in the promoter region of the ans gene (Fig. 2).
Two putative TATA boxes were found in the
Forsythia ans promoter (GenBank Y13435) at
po-sitions −84 and −97 from the first ATG codon.
No CAAT boxes were found upstream in the promoter. Sequence analysis revealed several
po-tential DNA – protein binding sites in the ansgene
promoter (Fig. 2). One of these motifs, the G box [19], was found in diverse gene promoters ([20], and Refs. therein). Conversely, other detected se-quences were reported to be binding sites for flavonoid pathway regulatory factors, namely: (a) one binding site for a MYB protein involved in the
petal epidermis-specific regulation of the Petunia
chsJ gene [21]; (b) six binding sites common to C1
and P, two MYB factors reported to regulate the maize anthocyanin and phlobaphene pathways, respectively [2,22,23]; (c) three sites for SBF-1, an
silencer/enhancer of a bean chs gene [24].
4. Discussion
This work deals with the molecular
characteri-zation of the ansin flower organs ofF.×interme
-dia cv. ‘Spring Glory’. The flavonoid pathway in
this widely grown variety has been investigated mainly to understand the block of anthocyanin synthesis in petals, in order to complement it by genetic transformation. Previous studies [6] had
already suggested a block ofansexpression as one
of the possible causes of the absence of an-thocyanins in petals. The results presented here
validated this hypothesis by showing that ans is
differentially expressed in Forsythia flower organs.
Data on 12 ANS polypeptide sequences are available to date, allowing improved sequence
analysis. Their length varies between 354
(Forsythia) and 430 (Petunia) residues, with the
Fig. 6. Northern analysis of ans expression in sepals of
Forsythia×intermedia cv. ‘Spring Glory’, using a FOANS1 – FOANS2 sepal cDNA probe. 0 – 4, flower developmental stages. Hybridization was carried out at 42°C in the presence of 50% formamide. Filter was finally washed in 0.1% SSPE, 0.1% SDS at 65°C for 15 min and exposed to X-ray films for 5 days.
containing the same 380-bp intron asF.×interme
-dia, was amplified from F. 6iridissima DNA. On
the other hand, two fragments of 969 and 876 bp,
containing introns of 536 bp (the same as F.×in
-termedia) and 443 bp, respectively, were amplified from F. suspensa var. fortunei.
3.3. Gene expression and ans promoter analyses
ans expression analysis was first accomplished
404-length of eight out of 12 between 354 (Forsythia)
and 362 (Vitis) amino acids. In spite of the
num-ber of sequences now available, information on
genomic organization of ans is limited to Vitis,
where it is a single copy gene [11]. InF.×interme
-dia cv. ‘Spring Glory’, Southern analysis
comple-mented by sequence data of genomic PCR
products indicated the presence of two ans alleles,
polymorphic for their introns. F.×intermedia has
been described as a natural hybrid between F.
suspensavar. fortunei andF. 6iridissima [25].
Nev-ertheless, this hypothesis has never been validated by genetic studies, hindering studies on putative
parents. Molecular analyses ofanssequences ofF.
suspensavar. fortunei and F.6iridissimasupported
the reported genealogy of F.×intermedia: F.
6iridissima provided the ‘short’ ans allele, and F.
suspensa the ‘long’ one to F.×intermedia.
Previous Northern analyses of ans expression
were reported forAntirrhinum[9],Petunia[10] and
Vitis [26]. In Forsythia, the use of PCR-based methods allowed fast and sensitive investigation of
ans expression in flower organs. The RT-PCR
results obtained with specific primers were confi-rmed with degenerate primers, thus scanning over
a potentialansfamily. This provided reliable
qual-itative information on organ-specific expression of
ansand indicated the lack ofanstranscript in both
anthers and petals. Northern blots indicate that
ansexpression in sepals has the same
developmen-tal pattern as that of chs in petals and — to a
lesser extent — that ofdfrin petals and sepals [6]:
the full dormancy stage 0 had the lowest ans
mRNA signal, and ans mRNA levels decreased
during flowering from stage 1 to 4. The similarity in above-mentioned gene expression patterns is in agreement with coordinated regulation of an-thocyanin pathway gene expression, already re-ported for model species [1,2]. By combining the
data ofans (this work) anddfr[6] gene expression
analyses in Forsythia, anthocyanin biosynthesis
was shown to be blocked in anthers at both thedfr
and ans levels and at the ans level in petals.
Studies on genetically defined genotypes or mu-tants showing blocks of anthocyanin pathway gene expression have already been reported in different plant species ([7,2], and Refs. therein). In
particular, Boss et al. [26] showed that in Vitisthe
absence of anthocyanins in skins of white berry
varieties is due to a block only of ufgt expression.
Therefore, it is possible that a single block of ans
expression could be the cause of the absence of
anthocyanin pigments in F.×intermedia cv.
‘Spring Glory’ petals. Transformation experiments
with a CaMV 35S promoter-driven M. incana ans
cDNA are underway in order to induce
an-thocyanin synthesis in Forsythia petals and test
this hypothesis.
The observed organ-specific expression of the
ans prompted ans promoter analyses. The
ana-lyzed ans promoter sequence should contain all
sequence sites required for regulation, as com-pared to other minimal anthocyanin promoters, less than 200 bp in length ([3], and Refs. therein). Besides the conserved G box, the presence of multiple binding sites for anthocyanin pathway-specific transcription factors is consistent with the model described in model species [2,3]. MYB.Ph3 factor belongs to the MYB class, one of the two families of regulatory factors necessary for activa-tion of anthocyanin genes [2,3]. As for C1 and P MYB factors, since anthocyanins are the only
flavonoid pigments in Forsythia flower organs [5],
it is likely that their putative binding sites would be recognized by a factor homologue to C1. C1 was shown to recognize diverse plant promoter sequences and has already been characterized ex-tensively ([3], and Refs. therein). SBF-1 was
re-ported to be a silencer/enhancer of a beanchsgene
[24], which has the same consensus binding site as GT-1, a transcription factor involved in the
light-dependent expression of a plantrbcS-3Agene [27].
In conclusion, characterization of ans pointed
out its critical role in the spatial and temporal
regulation of the anthocyanin pathway in
Forsythia flower organs.
Acknowledgements
We thank Professor G. Forkmann (Technische Universita¨t Mu¨nchen, Germany) for providing the
M. incana ans cDNA and sequence data.
References
[1] W. Heller, G. Forkmann, Biosynthesis of flavonoids, in: J.B. Harborne (Ed.), The Flavonoids: Advances in Re-search since 1986, Chapman and Hall, London, 1993, pp. 499 – 535.
[3] M.L. Lesnick, V.L. Chandler, Activation of the maize anthocyanin gene a2 is mediated by an element con-served in many anthocyanin promoters, Plant Physiol. 117 (1998) 437 – 445.
[4] J. Mol, E. Grotewold, R. Koes, How genes paint flowers and seeds, Trends Plant Sci. 3 (1998) 212 – 217.
[5] C. Rosati, A. Cadic, M. Duron, M.-J. Amiot, M. Tac-chini, S. Martens, G. Forkmann, Flavonoid metabolism in Forsythiaflowers, Plant Sci. 139 (1998) 133 – 140. [6] C. Rosati, A. Cadic, M. Duron, J.-P. Renou, P.
Si-moneau, Molecular cloning and expression analysis of dihydroflavonol 4-reductase gene in flower organs of
Forsythia×intermedia, Plant Mol. Biol. 35 (1997) 303 – 311.
[7] G. Forkmann, Precursors and genetic control of an-thocyanin synthesis in Matthiola incana R. Br, Planta 137 (1977) 159 – 163.
[8] W. Heller, L. Britsch, G. Forkmann, H. Grisebach, Leucoanthocyanidins as intermediates in anthocyanidin biosynthesis in flowers ofMatthiola incanaR. Br, Planta 163 (1985) 191 – 196.
[9] C. Martin, A. Prescott, S. Mackay, J. Bartlett, E. Vrij-landt, Control of anthocyanin biosynthesis in flowers of
Antirrhinum majus, Plant J. 1 (1991) 37 – 49.
[10] D. Weiss, A.H. van der Luit, J.T.M. Kroon, J.N.M. Mol, J.M. Kooter, The petunia homologue of theAntir
-rhinum majus candi and Zea mays A2 flavonoid genes shows homology to flavanone 3-hydroxylase and ethylene forming enzyme, Plant Mol. Biol. 22 (1993) 893 – 897.
[11] F. Sparvoli, C. Martin, A. Scienza, G. Gavazzi, C. Tonelli, Cloning and molecular analysis of structural genes involved in flavonoid and stilbene synthesis in grape (Vitis 6inifera L.), Plant Mol. Biol. 24 (1994) 743 – 755.
[12] A. Menssen, S. Hohmann, W. Martin, P.S. Schnable, P.A. Peterson, H. Saedler, A. Gierl, TheEn/Spm trans-posable element of Zea mayscontains splice sites at the termini generating a novel intron from dSpmelement in the A2 gene, EMBO J. 9 (1990) 3051 – 3057.
[13] K. Quandt, K. Frech, H. Karas, E. Wingender, T. Werner, MatInd and MatInspector — new fast and versatile tools for detection of consensus matches in nucleotide sequence data, Nucleic Acids Res. 23 (1995) 4878 – 4884.
[14] C. Rosati, A. Cadic, J.-P. Renou, M. Duron, Regenera-tion and Agrobacterium-mediated transformation of
Forsythia×intermedia‘Spring Glory’, Plant Cell Rep. 16 (1996) 114 – 117.
[15] J. Sambrook, E.F. Fritsch, T. Maniatis, Molecular Cloning: A Laboratory Manual, 2nd ed., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY,
1989.
[16] P.D. Siebert, A. Chenchik, D.E. Kellogg, K.A. Lukyanov, S.A. Lukyanov, An improved PCR method for walking in uncloned genomic DNA, Nucleic Acids Res. 23 (1995) 1087 – 1088.
[17] M.A. Frohman, M.K. Dush, G.R. Martin, Rapid pro-duction of full-length cDNAs from rare transcripts: am-plification using a single gene-specific oligonucleotide primer, Proc. Natl. Acad. Sci. USA 85 (1988) 8998 – 9002.
[18] A. Mahe, J. Grisvard, M. Dron, Fungal- and plant-spe-cific gene markers to follow the bean anthracnose infec-tion process and normalize a bean chitinase mRNA induction, Mol. Plant-Microbe Int. 5 (1992) 242 – 248. [19] G. Giuliano, E. Picherski, V.S. Malik, M.P. Timko, P.A.
Scolnik, A.R. Cashmore, An evolutionary conserved protein binding sequence upstream a plant light-regu-lated gene, Proc. Natl. Acad. Sci. USA 85 (1988) 7089 – 7093.
[20] R.G.K. Donald, A.R. Cashmore, Mutation of either G box or I box sequences profoundly affects expression from the Arabidopsis rbcS-1A promoter, EMBO J. 9 (1990) 1717 – 1726.
[21] R. Solano, C. Nieto, J. Avila, L. Can˜as, I. Diaz, J. Paz-Ares, Dual DNA binding specificity of a petal epi-dermis-specific MYB transcription factor (MYB.Ph3) fromPetunia hybrida, EMBO J. 14 (1995) 1773 – 1784. [22] E. Grotewold, B.J. Drummond, B. Bowen, T. Peterson,
Themyb-homologousPgene controls phlobaphene pig-mentation in maize floral organs by directly activating a flavonoid biosynthetic subset, Cell 76 (1994) 543 – 553. [23] M.B. Sainz, E. Grotewold, V.L. Chandler, Evidence for
direct activation of an anthocyanin promoter by the maize C1 protein and comparison of DNA binding by related Myb domain proteins, Plant Cell 9 (1997) 611 – 625.
[24] M.A. Lawton, S.M. Dean, M. Dron, J.M. Kooter, K.M. Kragh, M.J. Harrison, L. Yu, L. Tanguay, R.A. Dixon, C.J. Lamb, Silencer region of a chalcone synthase pro-moter contains multiple binding sites for a factor, SBF-1, closely related to GT-1, Plant Mol. Biol. 16 (1991) 235 – 249.
[25] D. Wyman, The Forsythia story, Arnoldia 21 (1961) 35 – 38.
[26] P.K. Boss, C. Davies, S.P. Robinson, Expression of anthocyanin biosynthesis pathway genes in red and white grapes, Plant Mol. Biol. 32 (1996) 565 – 569.
[27] P.J. Green, S.A. Kay, N.H. Chua, Sequence-specific interactions of a pea nuclear factor with light-responsive elements upstream of the rbcS-3A gene, EMBO J. 6 (1987) 2543 – 2549.