SA, JA, ethylene, and disease resistance in plants
Xinnian Dong
Exciting advances have been made during the past year: isolating mutants affecting plant disease resistance, cloning genes involved in the regulation of various defense responses, and characterizing novel defense signaling pathways. Recent studies have demonstrated that jasmonic acid and ethylene are important for the induction of nonspecific disease resistance through signaling pathways that are distinct from the classic systemic acquired resistance response pathway regulated by salicylic acid.
Addresses
Developmental, Cell, and Molecular Biology Group, Department of Botany, LSRC Building, PO Box 91000, Duke University, Durham, NC 27708-1000, USA; e-mail: [email protected]
Current Opinion in Plant Biology1998,1:316–323 http://biomednet.com/elecref/1369526600100316
Current Biology Ltd ISSN 1369-5266 Abbreviations
Avr avirulence
BTH benzo(1,2,3)thiadiazole-7-carbothioic acid S-methyl ester eds enhanced disease susceptibility
HR hypersensitive response INA 2,6-dichloroisonicotinic acid ISR induced systemic resistance JA jasmonic acid
MAP mitogen-activated protein
ndr non-race-specific disease resistance pad phytoalexin deficient
PR pathogenesis-related R resistance
SA salicylic acid
SAR systemic acquired resistance
Introduction
The battle between a plant and an invading pathogen is often dependent on speed. The winner is determined by how quickly the pathogen can proliferate and exert damage, compared to how fast the plant can respond with the necessary levels of defense. If a pathogen is immediately recognized by a plant, for example when a plant has a specific resistance (R) gene that interacts with the corresponding avirulence (avr) gene from the pathogen, a rapid defense mechanism known as the hypersensitive response (HR) occurs to prevent infection [1–6]. A lack of rapid pathogen recognition often leads to a successful infection. In addition to this immediate, one-on-one contest, plants also employ general resistance mechanisms induced after an HR or during a successful infection to combat secondary infections from a broad spectrum of pathogens or to prevent an existing infection from spreading further. One such general defense mech-anism is known as systemic acquired resistance (SAR) [7–9]. SAR induction requires the signal molecule salicylic acid (SA), which accumulates in plants prior to the onset
of SAR. Removal of SA in transgenic plants expressing salicylate hydroxylase (encoded by the bacterial nahG
gene) prevents the establishment of SAR [10]. In some plants such as tobacco, cucumber, and Arabidopsis, SA is not only necessary but also sufficient for the induction of SAR. Treatment of these plants with SA or its functional analogs such as 2,6-dichloroisonicotinic acid (INA) and benzo(1,2,3)thiadiazole-7-carbothioic acid S-methyl ester (BTH) induces SAR [11,12]. SAR is believed to be a result of the concerted activation of pathogenesis-related (PR) genes [13–16]; overexpression of a single PR gene only provides limited protection to the plant [17–21]. Even though their roles in disease resistance have yet to be clearly demonstrated, expression levels ofPRgenes serve as convenient markers for monitoring SAR.
Characterization of SAR in a variety of plant species has suggested the existence of a complex signaling network that involves many factors affecting various aspects of gen-eral disease resistance. It has become evident that plants utilize multiple pathways to transduce pathogenic signals to activate HR, SAR, and other resistance responses, and that SA-mediated SAR is not the only pathway that can lead to broad-spectrum disease resistance. Evidence is emerging that strongly suggests the importance of jasmonic acid (JA) and ethylene as alternative signals in the induction of resistance against microbial pathogens, in addition to their well-characterized roles in the wounding response in plants. In this review, I focus on the genetic and molecular experiments carried out in the past year that define the signaling components in both SA-dependent and SA-independent acquired resistance.
Different signaling pathways leading to the
induction of resistance
Genetic screens performed inArabidopsis have generated several classes of mutants which define potential signaling components downstream of the avr-R recognition event. One group of mutants, represented byeds1(for enhanced disease susceptibility) [22] andndr1(for non-race-specific disease resistance) [23], exhibit altered responses to multiple avr signals. The recessive eds1 mutation lacks the capability for signal transduction from a subset of R genes resulting in susceptibility to strains of
Peronospora parasiticaandPseudomonas syringaecarrying the corresponding avr genes. The ndr1 mutation appears to influence the function of a different subset of R genes causing a loss of resistance to Pseudomonas syringae pv.
tomato DC3000 carrying the bacterial avirulence genes
selective use of a signaling component may be based on the structure of an individual R gene, with each
R gene within a class sharing a particular component. Connections between these early signaling components and the downstream SA or JA and ethylene signaling pathways have yet to be established. The recent success in cloning the EDS1 gene which encodes a lipase (JE Parker, personal communication) and the NDR1 gene which encodes a protein with two putative transmembrane domains [24••] will certainly lead to new information on the molecular functions of these genes.
A new class of mutants, designated dnd(for defense, no death), do not show HR after inoculation with P. syringae
expressingavrRpt2oravrRpm1but maintain resistance to this pathogen (AF Bent, personal communication). The identification of dnd mutants may imply that cell death is not essential for avr–R specific resistance. Another plausible explanation is that the resistance observed in
dnd mutants is a result of the downstream activation of the SAR response, signified by the constitutively elevated levels of SA and PR gene expression in these mutant plants.
Analysis of mutants that form HR-like lesions in the absence of pathogen infection and display enhanced nonspecific pathogen resistance has yielded additional insights into regulation of resistance responses. The
Arabidopsis LSD1gene encodes a novel zinc finger protein which may respond to superoxide accumulated during the HR to restrict the spreading of cell death [25••]. The maize Lls1 gene has been found to encode a protein with two conserved substrate binding motifs of aromatic ring-hydroxylation dioxygenases and probably functions to degrade a phenolic mediator of cell death [26••]. The
Mlogene of barley is predicted to be a membrane bound protein with at least six membrane-spanning helices [27••]. Further characterization of these genes, in addition to the genes defined by the HR-compromised mutants, may provide clues as to how signals are produced during an HR leading to the induction of systemic resistance. Epistasis analyses between the HR-compromised mutants and the lesion mimic mutants may reveal the hierarchy of the signaling events.
A recent report has shown convincingly that reactive oxy-gen intermediates oxy-generated during an HR may serve as signals mediating the establishment of systemic resistance [28••]. Alvarez et al. demonstrated that inoculation of
Arabidopsis with an avirulent pathogen triggers not only an HR at the site of infection but also induces secondary oxidative bursts in discrete clusters of cells in distant, uninfected tissues, which ultimately lead to the formation of microscopic HR lesions. Both the primary oxidative burst and the reiterative secondary oxidative bursts in the systemic tissues have been shown to be required for the induction of SAR.
Virulent pathogens have also been used to identify components of nonspecific resistance responses. Char-acterization of these mutants has shown that there is a perception/signaling pathway for virulent pathogens leading to SA accumulation and resistance that is dis-tinct from that triggered by avirulent pathogens. The
eds mutants were isolated for their enhanced disease susceptibility after infection by the virulent pathogen
P. syringae pv. maculicola ES4326 [29,30••]. Inoculation of these mutant plants with an avirulent pathogen still results in an HR and an SAR response even though there is an upward shift in growth of the challenging pathogen in both control plants, pre-treated with 10 mM magnesium chloride, and induced plants, pre-inoculated with an avirulent pathogen. Ineds5-1,PR-1gene induction by the virulent pathogenPsmES4326 is reduced to 10% of the wild-type levels but is fully restored by SA treatment. Thepadmutants (for phytoalexin-deficient) are deficient in accumulating camalexin, an indolic phytoalexin, after infection by the virulent pathogen Psm ES4326 [31] and pad4, specifically, appears to affect a regulatory component in camalexin biosynthesis. The pad4 mutant displays enhanced susceptibility to Psm ES4326 and a number of isolates of P. parasitica [32•]. In addition, it exhibits pleiotropic defects in Psm ES4326-induced SA accumulation andPRgene expression that can be partially reversed by the addition of SA [33••]. The pleiotropic effects of pad4 are specific to the virulent pathogenPsm
ES4326 (and also Pst DC3000) because the synthesis of camalexin, the accumulation of SA, and the expression of
PR-1 gene induced by Psm ES4326 carrying avrRpt2are all similar to wild-type inpad4.
Characteristics of all these mutants suggest the existence of separate components involved in transducing diverse signals generated by different pathogens. PAD4 and EDS5 may be regulators controlled by signals produced during a virulent infection while NDR1, EDS1, DND and LSD1 may represent components involved in the
avr-induced HR, with NDR1 and EDS1 responding to distinct avr-Rinteractions. Without further knowledge of the signal molecules, however, classification of these signal transducing components can only be done arbitarily. A component identified in an avirulent pathogen-induced response, such as eds1, may also affect the response to a virulent pathogen. Nevertheless, all these components seem to be involved in early signaling events. Whether they play a direct role in the induction of SAR or other general resistance responses has yet to be determined.
SA-dependent signaling pathway
the resistance response is activated in these plants with constitutively high levels of SA. Using transgenic potato plants expressing bacterial salicylate hydroxylase (which metabolises SA), it has recently been demonstrated that SA is required for arachidonic acid-induced resistance to
Phytophthora infestansin potato. The resistance, however, is induced as a result of an increase in sensitivity to SA rather than an increase in SA synthesis [40••]. The mechanism of this induction is unknown; it could occur by increasing the accessibility of SA to a receptor, or by inducing synthesis or activity of an SA receptor.
Biochemical approaches used to identify components involved in the transduction of the SA signal have produced promising candidates. An SA binding protein has been identified [41•] which has a 150-fold higher affinity for SA than catalase: catalase being the first SA binding protein isolated using 14C-labeled SA. Protein
phosphorylation and dephosphorylation have been indi-cated in various defense responses including SA-regulated
PRgene expression [42]. Consistent with these results, a mitogen-activated protein (MAP) kinase has been shown to be activated in an SA-induced tobacco suspension cell extract [43••]. The sequence of this p48SIP kinase (SA-induced protein kinase) is distinct from other plant MAP kinases previously implicated in stress responses.
In addition to the biochemical approaches, genetic screens performed inArabidopsishave uncovered one locus,NPR1
(for nonexpresser of PR genes; also called NIM1 or
SAI1), that clearly functions downstream of SA [29,44–46].
NPR1 has been cloned independently by two research groups [47••,48••]. Sequence analysis of the gene and its twelve mutant alleles has shown that NPR1 contains a functionally important ankyrin-repeat domain which may be involved in protein–protein interaction. The carboxyl end of NPR1, where a nuclear localization signal (NLS) resides, has also been shown to be essential for protein function. The importance of this NLS has been demonstrated by the strict nuclear localization of a biologically active NPR1-GFP fusion protein after SAR induction (M Kinkema, X Dong, unpublished data). SAR induction not only modestly increases NPR1 levels, but also is required for activation of the NPR1 protein. Overexpression of NPR1 inArabidopsisconfers significant resistance to strains of P. syringae and P. parasitica that are normally virulent on wild-type Arabidopsis [49••]. By measuring the induction kinetics of thePRgenes during the infection, it has been demonstrated thatPRgenes are not constitutively expressed in the NPR1-overexpressing lines and that pathogen infections cause a stronger, but not faster, expression of these genes compared to wild-type. These characteristics make NPR1 a favorable target for genetic engineering of disease resistance in crops, because constitutive activation of SAR would be likely to have detrimental effects on plant growth and also might result in high selection pressure for more virulent pathogens. There are still many questions that
remain to be answered with regard to the molecular function of NPR1. For example, it is unknown if NPR1 regulates PR gene expression through activation of a positive transcription factor, or inactivation of a negative transcription factor. To completely understand NPR1 function, the NPR1-interacting component(s) will have to be identified and characterized.
JA- and ethylene-dependent signaling
pathway
An exciting new development in the past year is the discovery of SA-independent pathway(s) that also lead to broad-spectrum, systemic resistance. Both JA and ethylene have been shown to be important for the induction of these alternative responses. Fortunately, there already exists a wealth of information about the role of JA in plants response to wounding and insect attack [50]. The wound-induced octadecanoid pathway results in the synthesis of the signal molecule JA and subsequent activation of proteinase inhibitor genes. In addition to JA, ethylene has recently been shown to be a required, though not sufficient, signal that acts with JA to induce the wound response in tomato [51]. Ethylene is produced rapidly and transiently in leaves after injury or upon induction by JA or cell wall oligosaccharide fragments. Inhibition of ethylene production or sensitivity through mutation, antisense technology, or the use of chemical inhibitors all negatively affect induction of the JA pathway.
The JA regulated wound- and insect-induced pathway has long been thought to be involved in conferring resistance to microbial pathogens because it is also induced by oligosaccharide fragments released by the action of lytic enzymes produced by an invading pathogen; however, the role of ethylene and JA in plant disease resistance seemed at times to be ambiguous. For example, in the Arabidopsis ethylene-insensitive mutant ein2, disease symptoms caused by Pst DC3000, Psm 4326, and Xan-thomonas campestrispv.campestriswere shown to be reduced while growth of these pathogens remained unaffected [52,53]. In the same ethylene-insensitive mutant, the involvement of ethylene in SA-mediated SAR was ruled out because SA- and INA-induced PR-1gene expression and resistance toP. parasiticawere shown to be unaffected by the mutation [54]. In fact, the basal levels of PR-1
mRNA seemed to be higher in the ein2 mutant than in the wild-type. Similar results have recently been obtained using the tomato Never ripemutant impaired in ethylene perception [55•]. In another example, the Arabidopsis coi1
levels of PR gene expression and increased tolerance to bacterial pathogens.
A more clear-cut connection between systemic disease re-sistance, JA and ethylene signaling molecules has recently been established in Arabidopsisby the characterization of the systemically inducible antimicrobial peptides, thionin Thi2.1 and defensin PDF1.2 [57,58]. The expression of Thi2.1 is induced by methyl JA, silver nitrate, and
Fusarium oxysporum f. sp. matthiolae, but not by SA [57]. The antimicrobial activity of thionin has been demonstrated in Arabidopsis by the enhanced resistance toF. oxysporumspmatthiolaeobserved in transgenic plants overexpressing theThi2.1gene [59•]. ThePDF1.2gene is induced by JA, ethylene, rose bengal, and the non-host pathogen Alternaria brassicicola [58]. The induction of
PDF1.2byA. brassicicolais inhibited in the JA-insensitive mutant coi1and in the ethylene-insensitive mutant ein2, indicating that JA and ethylene are required for the induction. PDF1.2 expression, however, is unaffected in
nahG transgenic plants which are unable to accumulate SA, in the npr1 mutant which is insensitive to SA induction, or in thecpr1mutant which has elevated levels of SA and constitutive SAR. It is evident that PDF1.2 and Thi2.1 represent end products of a systemically expressed resistance pathway(s) that is distinct from the SA-dependent SAR and is potentially regulated by both JA and ethylene.
Genetic studies have shown that JA and ethylene are essential signals in another resistance response known as induced systemic resistance (ISR) that is established after treatment with various strains of root-colonizing biocontrol bacteria Pseudomonas fluorescens (CMJ Pieterse, personal communication). ISR is effective against the virulent Pst DC3000 and F. oxysporumf. sp. raphani and functions independently of SA and PR gene expression [60,61•]. Intriguingly, during ISR,PDF1.2is not induced, suggesting that JA and ethylene may regulate ISR through a pathway different from that which includes PDF1.2 and Thi2.1. ISR, therefore, appears to be a novel systemic resistance response in plants that can be triggered in roots by rhizobacteria and is regulated by JA and ethylene.
Interactions between different resistant
pathways
The antagonistic relationship between the JA and SA pathways has been well documented in the studies of wound responses in plants [50,62]. The inhibitory effect of SA on the proteinase inhibitor genes is partially overcome by pretreatment of plants with both JA and ethylene, but not by either signal alone [51]. This further supports the notion that both ethylene and JA are required signals for wound responses and implies that SA inhibits not only the synthesis of JA and ethylene but also a signaling step downstream of JA and ethylene.
Interactions between the different pathways are also evi-dent in plant resistance responses to microbial organisms.
For example, in tobacco, treatments with SA and the pathogenicErwinia carotovoraorErwinia-derived elicitors seem to induce two distinct pathways leading to the expression of separate sets of genes [63•].Erwinia-derived elicitors antagonize the SA-mediated induction of PR
genes while SA inhibits the induction ofErwinia-activated genes. Such antagonism suggests the presence of common regulatory components for both pathways, and possibly signifies the evolution of a mechanism designated to prioritize different resistance responses.
Common regulatory components have been found in sepa-rate signaling pathways. In the root-colonizing bacterium-induced ISR, which is SA-independent but JA- and ethylene-dependent, the function of NPR1 has been shown to be essential (CMJ Pieterse, personal com-munication). This implies that NPR1 not only plays a key regulatory role in SAR but also participates in the JA- and ethylene-regulated, SA-independent ISR. This is consistent with the detection of high levels of NPR1 in roots (H Cao, M Kinkema, X Dong, unpublished data) where ISR is induced and wherePRgene expression and SAR are absent [64].
Other components potentially shared by the SA-induced SAR and the JA- and ethylene-induced disease resistance pathways have been identified through epistasis analyses betweenArabidopsismutants with enhanced resistance and mutants with compromised defense responses. The cpr5
mutant expresses both the PR genes and the PDF1.2,
and Thi2.1 genes, and is resistant to Psm ES4326 and
P. parasitica Noco2. In the cpr5:npr1 double mutant, the expression ofPRgenes and resistance toPsmES4326 are abolished while the expression of PDF1.2 and resistance to P. parasitica remain unaffected [65••]. These results suggest that a mutation in a single gene can upregulate an NPR1-dependent pathway and an NPR1-independent pathway, both of which can provide resistance to P. parasitica. This conclusion is supported by the epistasis analysis performed in thecpr1:npr1double mutant. Incpr1, where only the NPR1-dependent pathway is activated, the enhanced resistance toP. parasiticais completely abolished by npr1in the cpr1:npr1 double mutant (SA Bowling, X Dong, unpublished data).
Analyses of the dominant, gain-of-resistance Arabidopsis
mutant cpr6 define another possible point of interaction between the SA-regulated and the JA- and ethylene-reg-ulated pathways [66••]. Similar to cpr5, the cpr6 mutant displays enhanced levels ofPRandPDF1.2gene expres-sion. In the cpr6:npr1 double mutant, however, neither
PR nor PDF1.2 gene expression is significantly affected by npr1. Even though it is difficult to determine the precise position ofcpr6in the disease resistance signaling network, the data suggest a dual role forcpr6in regulating both the PR and PDF1.2 genes. Perhaps the wild-type CPR6 works with NPR1 to induce the expression of
the protein NPR1-independent. The interplay between CPR6, NPR1 and the two distinct signaling pathways has been further suggested by the complete suppression of
PDF1.2 expression by INA treatment observed in cpr6, in comparison to only a partial suppression of PDF1.2
expression in the cpr6:npr1 double mutant. A functional NPR1 protein seems to be required for INA to sequester all the mutant cpr6 protein thereby preventing activation ofPDF1.2.
The relationship between the SA-regulated SAR and the JA- and ethylene-regulated disease resistance needs to be explored further using mutants of both pathways. For example, epistasis analysis between the cpr5 or cpr6
mutant and the ethylene-insensitive mutant, ein2, or the jasmonic acid-non-responsive mutant, jar1 [67], will be carried out and the results will be compared with those of
cpr5:npr1orcpr6:npr1. Preliminary analysis of thecpr5:ein2
and cpr6:ein2 double mutants have indicated that a full
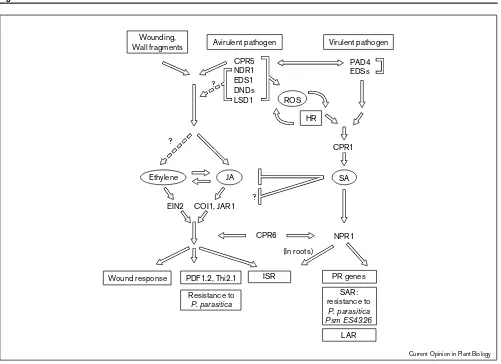
Figure 1
Ethylene
CPR5 NDR1 EDS1 DNDs LSD1
PAD4 EDSs
CPR1
SA JA
COI1, JAR1 EIN2
NPR1 CPR6
(In roots) ?
?
ISR PR genes
LAR SAR: resistance to
P. parasitica Psm ES4326
?
HR ROS Wounding,
Wall fragments
Wound response PDF1.2, Thi2.1 Resistance to
P. parasitica
Avirulent pathogen Virulent pathogen
Current Opinion in Plant Biology
A working model of pathways leading to broad-spectrum disease resistance in plants. TheArabidopsisgenes defined by the mutants described in this review are presented. After plant–pathogen recognition, multiple signaling components (CPR5, NDR1, EDS1, DNDs, LSD1) are employed that lead to the formation of reactive oxygen species (ROS) and HR, and the synthesis of SA. NDR1 and EDS1 are involved in transducing signals from separate subsets ofavr-Rinteractions, the relative positions of other genes have yet to be determined. It is also unknown whether these genes (exceptCPR5) are involved in the JA- and ethylene-mediated resistant responses. (This ambiguity is represented by a dashed line.) Virulent pathogens can also induce SA synthesis and local resistance, probably through separate regulatory components such as PAD4 and EDSs. Further downstream, the CPR1-SA-NPR1 linear pathway has been demonstrated by various genetic analyses. The pathway of JA and ethylene is proposed on the basis of the wound response in tomato [51], showing that JA and ethylene induce each other’s synthesis and that JA and ethylene are both required for activating the wound response,PDF1.2andThi2.1gene expression, resistance toP. parasitica, and ISR. It remains to be determined whether these responses (shown in rectangular boxes) share regulatory components downstream of JA and ethylene. The SA pathway and the JA/ethylene pathway may interact antagonistically, with SA inhibiting both the synthesis and the signal transduction of JA and ethylene (illustrated with blocked lines). The possible involvement of NPR1 in the SA-independent but JA- and ethylene-dependent ISR is intriguing. Further characterization of NPR1 and anotherArabidopsiscomponent CPR6, which is involved in regulating the expression of both
induction ofPDF1.2incpr5andcpr6requires the function of EIN2 and probably ethylene (JD Clarke, X Dong, unpublished data).
Conclusions
Isolation of different classes of mutants affecting nonspe-cific disease resistance and characterization of various plant defense responses have revealed a complex and interesting network of pathways that function both separately and together to render broad-spectrum protection to plants. At the center of this network are the signal molecules SA, JA, and ethylene. A model, with the Arabidopsis
genes described in this review highlighted, is represented here to stimulate more discussion (Figure 1). After plant–pathogen recognition, multiple signaling compo-nents (CPR5, NDR1, EDS1, DNDs, LSD1) are employed that lead to the formation of reactive oxygen species (ROS) and HR, and the synthesis of SA. While NDR1 and EDS1 are involved in transducing signals from separate subsets ofavr-Rinteractions, the relative positions of other genes have yet to be determined. It is also unknown whether these genes (except CPR5) are involved in the JA- and ethylene-mediated resistant responses. Virulent pathogens can also induce SA synthesis and local resistance, probably through separate regulatory components such as PAD4 and EDSs. Further downstream, the CPR1-SA-NPR1 linear pathway has been demonstrated by various genetic analyses. The pathway of JA and ethylene is proposed on the basis of the wound response in tomato [51] showing that JA and ethylene induce each other’s synthesis and that JA and ethylene are both required for activating the wound response, PDF1.2 and Thi2.1 gene expression, resistance to P. parasitica, and ISR. It remains to be determined whether these responses share regulatory components downstream of JA and ethylene. The SA pathway and the JA/ethylene pathway may interact antagonistically, with SA inhibiting both the synthesis and the signal transduction of JA and ethylene. The possible involvement of NPR1 in the SA-independent but JA- and ethylene-dependent ISR is intriguing. Further characterization of NPR1 and another Arabidopsis component CPR6, which is involved in regulating the expression of bothPRandPDF1.2genes, should provide new insight into the relationship of these pathways.
A complete understanding of these signaling events calls for co-operation between laboratories working on different resistance responses. As more epistasis tests are carried out between different classes of mutants, new resistance-related phenotypes will be revealed and used to perform more in-depth analyses. Information from both genetic and molecular research will be combined to establish a more comprehensive outline of the disease resistance network in plants.
Acknowledgments
I would like to thank AF Bent, J Glazebrook, JE Parker, CMJ Pieterse, AD Shapiro for communicating unpublished work and JD Clarke, J Glazebrook, M Kinkema, JN Siedow for helpful suggestions on this review.
References and recommended reading
Papers of particular interest, published within the annual period of review have highlighted as:
• of special interest
•• of outstanding interest
1. Flor HH:Current status of gene-for-gene concept.Annu Rev Phytopathol1971,9:275-296.
2. Hammond-Kosack K, Jones, JDG:Resistance gene-dependent plant defense responses.Plant Cell1996,8:1773-1791. 3. Keen NT:Gene-for-gene complementarity in plant–pathogen
interactions.Annu Rev Genet1992,24:447-463.
4. Lamb CJ:Plant disease resistance genes in signal perception and transduction.Cell1994,76:419-422.
5. Lamb CJ, Lawton MA, Dron M, Dixon RA:Signals and transduction mechanisms for activation of plant defenses against microbial attack.Cell1989,56:215-224.
6. Staskawicz BJ, Ausubel FM, Baker BJ, Ellis JG, Jones JDG: Molecular genetics of plant disease resistance.Science1995, 268:661-667.
7. Kuc J:Induced immunity to plant disease.BioScience1982, 32:854-860.
8. Ross AF:Systemic acquired resistance induced by localized virus infections in plants.Virology1961,14:340-358. 9. Ryals JA, Neuenschwander UH, Willits MG, Molina A, Steiner
H-Y, Hunt MD:Systemic acquired resistance.Plant Cell1996, 8:1809-1819.
10. Gaffney T, Friedrich L, Vernooij B, Negrotto D, Nye G, Uknes S, Ward E, Kessmann H, Ryals J:Requirement of salicylic acid for the induction of systemic acquired resistance.Science1993, 261:754-756.
11. M ´etraux J-P, Ahl-Goy P, Staub T, Speich J, Steinemann A, Ryals J, Ward E:Induced resistance in cucumber in response to 2,6-dichloroisonicotinic acid and pathogens.InAdvances in Molecular Genetics of Plant-Microbe Interactions 1Edited by Hennecke H, Verma DPS. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1991:432-439.
12. G ¨orlach J, Volrath S, Knauf-Beiter G, Hengy G, Beckhove U, Kogel K-H, Oostendorp M, Staub T, Ward E, Kessmann H, Ryals J: Benzothiadiazole, a novel class of inducers of systemic acquired resistance, activates gene expression and disease resistance in wheat.Plant Cell1996,8:629-643.
13. Uknes S, Mauch-Mani B, Moyer M, Potter S, Williams S, Dincher S, Chandler D, Slusarenko A, Ward E, Ryals J:Acquired resistance inArabidopsis.Plant Cell1992,4:645-656. 14. Van Loon LC, Van Kammen A:Polyacrylamide disc
electrophoresis of the soluble proteins fromNicotiana tabacumvar. Samsun and Samsun NN. II. Changes in protein constitution after infection with tobacco mosaic virus.Virology
1970,40:199-211.
15. Ward ER, Uknes SJ, Williams SC, Dincher SS, Wiederhold DL, Alexander DC, Ahl-Goy P, M ´etraux J-P, Ryals JA:Co-ordinate gene activity in response to agents that induce systemic acquired resistance.Plant Cell1991,3:1085-1094.
16. Yalpani N, Silverman P, Wilson TMA, Kleier DA, Raskin I:Salicylic acid is a systemic signal and an inducer of pathogenesis-related proteins in virus-infected tobacco.Plant Cell1991, 3:809-818.
17. Alexander D, Glascock C, Pear J, Stinson J, Ahl-Goy P, Gut-Rella M, Goodman RM, Ryals J:Systemic acquired resistance in tobacco: use of transgenic expression to study the functions of pathogenesis-related proteins.InAdvances in Molecular Genetics of Plant–Microbe Interactions.Edited by Nester EW, Verma DPS. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1993:527-533.
18. Alexander D, Goodman RM, Gut-Rella M, Glascock C, Weymann K, Friedrich L, Maddox D, Ahl-Goy P, Luntz T, Ward E, Ryals J: Increased tolerance to two Oomycete pathogens in transgenic tobacco expressing pathogenesis-related protein 1a.Proc Natl Acad Sci USA1993,90:7327-7331.
resistance to the fungal pathogenRhizoctonia solani.Science
1991,254:1194-1197.
20. Liu D, Kashchandra G, Raghothama P, Hasegawa PM, Bressan RA:Osmotin overexpression in potato delays development of disease symptoms.Proc Natl Acad Sci USA1994, 91:1888-1892.
21. Zhu Q, Maher EA, Masoud S, Dixon RA, Lamb CJ:Enhanced protection against fungal attack by constitutive coexpression of chitinase and glucanase genes in transgenic tobacco.
Bio/Technology1994,12:807-812.
22. Parker JE, Holub EB, Frost LN, Falk A, Gunn ND, Daniels MJ:Characterization ofeds1, a mutation inArabidopsis
suppressing resistance toPeronospora parasiticaspecified by several differentRPPgenes.Plant Cell1996,8:2033-2046. 23. Century KS, Holub EB, Staskawicz BJ:NDR1, a locus of
Arabidopsis thalianathat is required for disease resistance to both a bacterial and a fungal pathogen.Proc Natl Acad Sci USA1995,92:6597-6601.
••
24. Century KS, Shapiro AD, Repetti PP, Dahlbeck D, Holub E, Staskawicz BJ:NDR1, a pathogen-induced component required forArabidopsisdisease resistance.Science1998, 278:1963-1965.
The authors describe positional cloning of the Arabidopsis NDR1 gene which is required for the expression of resistance against both bacterial and fungal pathogens mediated by severalRgene products. The NDR1 protein contains two putative transmembrane domains.
••
25. Dietrich RA, Richberg MH, Schmidt R, Dean C, Dangl JL:A novel zinc finger protein is encoded by theArabidopsis LSD1gene and functions as a negative regulator of plant cell death.Cell
1997,88:685-694.
The authors report positional cloning of theArabidopsis LSD1gene. The molecular function of LSD1 as a negative regulator of cell death in plants is suggested on the basis of amino acid sequence information which predicts the protein contains three zinc finger domains.
••
26. Gray J, Close PS, Briggs SP, Johal GS:A novel suppressor of cell death in plants encoded by theLls1gene of maize.Cell
1997,89:25-31.
The maizeLls1gene cloned by transposon-tagging encodes a protein with two aromatic ring-hydroxylating dioxygenase substrate binding motifs. It is suggested to play a role in controlling the spread of cell death in mature maize leaves by degrading a phenolic mediator of cell death.
••
27. B ¨uschges R, Hollricher K, Panstruga R, Simons G, Wolter M, Frijters A, van Daelen R, van der Lee T, Diergaarde P, Groenendijk Jet al.:The barleyMlogene: A novel control element of plant pathogen resistance.Cell1997,88:695-705. The authors report positional cloning of the barleyMlogene which, when mutated, causes leaf lesioning and broad spectrum resistance toErysiphe graminisf. sp.hordei. TheMlogene is shown to encode a protein with at least six membrane-spanning helices, and its dual function in down regulating cell death and resistance is discussed.
••
28. Alvarez ME, Pennell RI, Meijer PJ, Ishikawa A, Dixon RA, Lamb C: Reactive oxygen intermediates mediate a systemic signal network in the establishment of plant immunity.Cell1998, 92:773-784.
The authors show that inoculation ofArabidopsiswith an avirulent pathogen not only leads to an immediate oxidative burst at the site of infection but also result in reiterative oxidative bursts in the uninfected systemic tissues, which eventually lead to formation of microscopic HR. Both the primary and the secondary oxidative bursts are required for the establishment of SAR. 29. Glazebrook J, Rogers EE, Ausubel FM:Isolation ofArabidopsis
mutants with enhanced disease susceptibility by direct screening.Genetics1996,143:973-982.
••
30. Rogers EE, Ausubel FM:Arabidopsisenhanced disease susceptibility mutants exhibit enhanced susceptibility to several bacterial pathogens and alterations inPR-1gene expression.Plant Cell1997,9:305-316.
An in-depth characterization of previously isolated Arabidopsis mutants
eds5-1, eds6-1, eds7-1 andeds9-1which have enhanced disease sus-ceptibility to a virulent bacterial pathogen. Each of the mutants shows a distinguishable phenotype in response to a panel of bacterial pathogens and with regard toPR-1gene inducibility. The data suggest the presence of an unknown resistance mechanism against virulent pathogens.
31. Glazebrook J, Ausubel FM:Isolation of phytoalexin-deficient mutants ofArabidopsis thalianaand characterization of their interactions with bacterial pathogens.Proc Natl Acad Sci USA
1994,91:8955-8959.
•
32. Glazebrook J, Zook M, Mert F, Kagan I, Rogers EE, Crute IR, Holub EB, Hammerschmidt R, Ausubel FM:Phytoalexin-deficient
mutants ofArabidopsisreveal thatPAD4encodes a regulatory factor and that fourPADgenes contribute to downy mildew resistance.Genetics1997,146:381-392.
Among the fivepadmutants (phytoalexin deficient) described in this paper,
pad4is shown to affect the regulation of camalexin biosynthesis after in-fection byPseudomonas syringae, but the mutant is still able to produce camalexin whenCochliobolus carbonumis used in the infection.pad4not only shows enhanced disease susceptibility toPseudomonas syringaebut also is fully susceptible to four of the six incompatible strains ofPeronospora parasiticatested.
••
33. Zhou N, Tootle TL, Tsui F, Klessig DF, Glazebrook J:PAD4
functions upstream from salicylic acid to control defense responses inArabidopsis.Plant Cell1998, in press.
An in-depth characterization of the previously isolatedpad4mutant. pad4
is shown to have pleiotropic defects on camalexin synthesis andPR-1gene expression after infection by the virulent pathogenPseudomonas syringae
pv.maculicolaES4326. SA synthesis induced byPsmES4326 is also re-duced and delayed inpad4. Treatment ofpad4with SA partially restores camalexin synthesis andPRgene expression. Therefore,PAD4is speculated to act upstream of SA accumulation in response toPsmES4326 infection. A model is proposed to illustrate the function of PAD4.
34. Malamy J, Carr JP, Klessig DF, Raskin I:Salicylic acid: A likely endogenous signal in the resistance response of tobacco to viral infection.Science1990,250:1002-1004.
35. M ´etraux J-P, Signer H, Ryals J, Ward E, Wyss-Benz M, Gaudin J, Raschdorf K, Schmid E, Blum W, Inverardi B:Increase in salicylic acid at the onset of systemic acquired resistance in cucumber.
Science1990,250:1004-1006.
36. Uknes S, Winter A, Delaney T, Vernooij B, Morse A, Friedrich L, Nye G, Potter S, Ward E, Ryals J: Biological induction of systemic acquired resistance inArabidopsis.Mol Plant-Microbe Interact1993,6:692-698.
37. Silverman P, Seskar M, Kanter D, Schweizer P, M ´etraux J-P, Raskin I:Salicylic acid in rice: biosynthesis, conjugation and possible role.Plant Physiol1995,108:633-639.
•
38. Chen Z, Iyer S, Caplan A, Klessig DF, Fan B:Differential accumulation of salicylic acid and salicylic acid-sensitive catalase in different rice tissues.Plant Physiol1997, 114:193-201.
In this report, the authors argue that SA regulates a defense response in rice through differential accumulation of SA and tissue-specific expression of catalases with different levels of sensitivities to SA.
39. Coquoz JL, Buchala AJ, Meuwly PH, M ´etraux J-P:Arachidonic acid induces local but not systemic synthesis of salicylic acid and confers systemic resistance in potato plants to
Phytophthora infestansandAlternaria solani.Phytopathology
1995,85:1219-1224.
••
40. Yu D, Liu Y, Fan B, Klessig DF, Chen Z:Is the high basal level of salicylic acid important for disease resistance in potato?Plant Physiol1997,115:343-349.
To investigate the role of SA in controlling resistance in potato plants where high basal levels of SA are detected, the bacterial salicylate hydroxylase (nahG) gene was introduced into potato. The resulting transgenic plants show no significant increase in disease severity when infected by Phytoph-thora infestans, while the arachidonic acid-induced systemic resistance to the same pathogen is abolished. These results indicate that SA is a nec-essary, but not sufficient, signal for arachidonic acid-induced resistance in potato and the development of SAR may involve an increase in sensitivity to SA.
•
41. Du H, Klessig DF: Identification of a soluble, high-affinity salicylic acid-binding protein in tobacco.Plant Physiol1997, 113:1319-1327.
The authors describe the identification of a new SA-binding, soluble protein with a low abundance in tobacco leaves and a 150-fold higher affinity for SA than that shown by catalase.
42. Conrath U, Silva H, Klessig DF:Protein dephosphorylation mediates salicylic acid-induced expression ofPR-1genes in tobacco.Plant J1997,11:747-757.
••
43. Zhang S, Klessig DF: Salicylic acid activates a 48 kD MAP kinase in tobacco.Plant Cell1997,9:809-824.
Using an in-gel kinase assay, a MAP kinase is shown to be activated rapidly and transiently in tobacco suspension cells after addition of SA. The protein has been purified, and the corresponding gene cloned and shown to be distinct from other previously identified stress-induced MAP kinases. 44. Cao H, Bowling SA, Gordon AS, Dong X:Characterization of
45. Delaney TP, Friedrich L, Ryals JA:Arabidopsissignal transduction mutant defective in chemically and biologically induced disease resistance.Proc Natl Acad Sci USA1995, 92:6602-6606.
46. Shah J, Tsui F, Klessig DF:Characterization of a salicylic acid-insensitive mutant (sai1) ofArabidopsis thaliana, identified in a selective screen utilizing the SA-inducible expression of the
tms2gene.Mol Plant–Microbe Interact1997,10:69-78.
••
47. Cao H, Glazebrook J, Clarke JD, Volko S, Dong X:The
Arabidopsis NPR1gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats.
Cell1997,88:57-63.
The authors describe positional cloning of theArabidopsis NPR1gene and analysis of fournpr1mutant alleles. The functional importance of the ankyrin-repeat domain found in NPR1 is speculated upon. Transforming thenpr1
mutants with the wild-type NPR1 genomic clone not only complements the mutant phenotypes, but also results in enhanced resistance.
••
48. Ryals J, Weymann K, Lawton K, Friedrich L, Ellis D, Steiner J-Y, Johnson J, Delaney TP, Jesse T, Vos P, Uknes S:TheArabidopsis NIM1protein shows homology to the mammalian transcription factor inhibitor IB.Plant Cell1997,9:425-439.
The authors describe positional cloning of theArabidopsis NIM1gene and characterization of five newnim1mutant alleles. Sequence homology be-tween NIM1 and the mammalian transcription factor inhibitor IB is outlined and the possible implication of this finding is discussed.
••
49. Cao H, Li X, Dong X:Generation of broad-spectrum disease resistance by overexpression of an essential regulatory gene in systemic acquired resistance.Proc Natl Acad Sci USA1998, in press.
The authors present results which prove the concept that broad-spectrum disease resistance can be generated through manipulation of theNPR1
gene, a regulatory of gene of SAR. NPR1 is shown to determine resistance in a dosage-dependent fashion. Overexpression of NPR1 leads to significant resistance to virulent strains ofPseudomonas syringaeandPeronospora parasiticawith little detrimental effect on the growth of the plants. 50. Doares SH, Syrovets T, Weiler EW, Ryan CA:
Oligogalactur-onides and chitosan activate plant defensive genes through the octadecanoid pathway.Proc Natl Acad Sci USA1995, 93:4095-4098.
51. ODonnell PJ, Calvert C, Atzorn R, Wasternack C, Leyser HMO, Bowles DJ:Ethylene as a signal mediating the wound response of tomato plants.Science1996,274:1914-1917. 52. Guzm ´an P, Ecker JR:Exploiting the triple response of
Arabidopsisto identify ethylene-related mutants.Plant Cell
1990,2:513-523.
53. Bent AF, Innes RW, Ecker JR, Staskawicz BJ:Disease development in ethylene-insensitiveArabidopsis thaliana
infected with virulent and avirulentPseudomonasand
Xanthomonaspathogens.Mol Plant–Microbe Interact1992, 5:372-378.
54. Lawton KA, Potter SL, Uknes S, Ryals J:Acquired resistance signal transduction inArabidopsisis ethylene independent.
Plant Cell1994,6:581-588.
•
55. Lund ST, Stall RE, Klee H:Ethylene regulates the susceptible response to pathogen infection in tomato.Plant Cell1998, 10:371-382.
Using the tomato Never ripemutant impaired in ethylene perception, the authors show that development of disease symptoms caused by the virulent pathogensXanthomonas campestrispv.vesicatoria, Pseudomonas syringae
pv.tomatoandFusarium oxysporumf. sp.lycopersicirequires ethylene. In the mutant, the disease symptoms are reduced compared to wild-type, but growth of the pathogens remains unaffected.
56. Feys BJF, Benedetti CE, Penfold CN, Turner JG:Arabidopsis
mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate, and show resistance to a bacterial pathogen.Plant Cell1994,6:751-759.
57. Epple P, Apel K, Bohlmann H:AnArabidopsis thalianathionin gene is inducible via a signal transduction pathway different from that for pathogenesis-related proteins.Plant Physiol1995, 109:813-820.
58. Penninckx IAMA, Eggermont K, Terras FRG, Thomma BPHJ, De Samlanx GW, Buchala A, M ´etraux J-P, Manners J, Broekaert WF: Pathogen-induced systemic activation of a plant defensin gene inArabidopsisfollows a salicylic acid-independent pathway.
Plant Cell1996,8:2309-2323.
•
59. Epple P, Apel K, Bohlmann H:Overexpression of an endogenous thionin enhances resistance ofArabidopsis
againstFusarium oxysporum.Plant Cell1997,9:509-520. The authors demonstrate the antimicrobial activity of thionin by overexpress-ing an endogenous thionin geneThi2.1inArabidopsis. The transgenic line is shown to have enhanced resistance againstFusarium oxysporum. 60. Pieterse CMJ, van Wees SCM, Hoffland E, van Pelt JA, van Loon
LC:Systemic resistance inArabidopsisinduced by biocontrol bacteria is independent of salicylic acid accumulation and pathogenesis-related gene expression.Plant Cell1996, 8:1225-1237.
•
61. Van Wees SCM, Pieterse CMJ, Trijssenaar A, Vant Westende YAM, Hartog F, Van Loon LC:Differential induction of systemic resistance inArabidopsisby biocontrol bacteria.Mol Plant Microbe–Interact1997,10:716-724.
The authors show that induced systemic resistance (ISR) triggered in Ara-bidopsisby biocontrol bacteria involves a specific interaction between the plant and the pathogen, which is reflected in the ecotype- and strain-speci-ficity observed in ISR. Mutant bacteria lacking the O-antigenic side chain of lipopolysaccharide (LPS) are still effective in inducing ISR, suggesting that in addition to LPS, other factors are also involved in triggering ISR in
Arabidopsis.
62. Doares SH, Narvaez-Vasquez J, Conconi A, Ryan CA:Salicylic acid inhibits synthesis of proteinase inhibitors in tomato leaves induced by systemin and jasmonic acid.Plant Physiol
1995,108:1741-1746.
•
63. Vidal S, de Leon IP, Denecke J, Palva ET:Salicylic acid and the plant pathogenErwinia carotovorainduce defense genes via antagonistic pathways.Plant J1997,11:115-123.
UsingErwiniaand SA as inducers, the authors describe two distinct path-ways leading to expression of separate sets of genes. SA- and Erwinia-de-rived signals have antagonistic effects on each other.
64. Bowling SA, Guo A, Cao H, Gordon AS, Klessig DF, Dong X:A mutation inArabidopsisthat leads to constitutive expression of systemic acquired resistance.Plant Cell1994,6:1845-1857.
••
65. Bowling SA, Clarke JD, Liu Y, Klessig DF, Dong X:Thecpr5
mutant ofArabidopsisexpresses both NPR1-dependent and NPR1-independent resistance.Plant Cell1997,9:1573-1584. The authors report the results of an epistasis analysis between the Arabidop-sis cpr5mutant (with enhanced resistance) and thenpr1mutant (with im-paired SAR). An NPR1-independent pathway is uncovered incpr5which is shown to confer resistance to the virulent pathogenPeronospora parasitica. A model is presented to illustrate the NPR1-dependent and -independent pathways.
••
66. Clarke JD, Liu Y, Klessig DF, Dong X:Uncoupling PR gene expression from NPR1 and bacterial resistance: Characterization of the dominantArabidopsis cpr6-1mutant.
Plant Cell1998,10:557-569.
Through genetic analyses of the dominantArabidopsis cpr6mutant which has constitutive expression of both the SA-regulated PRgenes and the JA- and ethylene-regulatedPDF1.2gene, the authors show that CPR6 is involved in multiple resistance pathways, and thatPRgene expression does not confer resistance to the bacterial pathogenPseudomonas syringae ob-served in thecpr6mutant.