The ABC model of flower development represents a milestone in explaining how the fate of emerging floral organ primordia is specified. This model states that organ identity is specified by different combinations of the activities of the A, B and C class homeotic genes. In spite of the remarkable simplicity of this model, the complex regulatory interactions that establish the initial pattern of A, B and C gene activity have yet to be fully explained. It has been shown that the LEAFYgene functions early to promote flower meristem identity, and that it is subsequently required for the normal expression of the ABC genes. Recently, LEAFYhas been identified as an immediate upstream regulator of the floral homeotic genes, thus opening up an avenue to examine the transcriptional interactions that underlie floral patterning.
Addresses
Department of Biology, University of California at San Diego, La Jolla, California, 92093-0116, USA
*e-mail: [email protected]
†e-mail: [email protected]
Current Opinion in Plant Biology2000, 3:47–52
1369-5266/00/$ — see front matter © 2000 Elsevier Science Ltd. All rights reserved.
Abbreviations
AG AGAMOUS
AP APETALA
FIM FIMBRIATA
LFY LEAFY
PI PISTILLATA
SAM shoot apical meristem
UFO UNUSUAL FLORAL ORGANS
Introduction
The Arabidopsis flower is arguably the best understood plant model system pertaining to pattern formation. Flowers originate from small groups of undifferentiated cells called floral meristems, which, in turn, are derived from the shoot apical meristem (SAM). Similar to most other dicotyledonous plants, an Arabidopsis flower consists of four types of floral organs arranged in four concentric whorls. Four sepals, four petals, six stamens and two fused carpels can be found from the periphery to the center of each flower.
In order to understand how flowers develop into their final shape and form, a genetic approach has been used to iden-tify genes that when inactivated will perturb the flower morphology. In this review, we focus on the meristem iden-tity genes and the floral homeotic genes. Analyses of mutations in the latter class of genes led to the formulation of the ‘ABC’ model of flower development (reviewed in [1,2]). According to this model, the A genes specify sepals, the A and B genes together specify petals, the B and C genes together specify stamens, and the C gene specifies
carpels. An example of the A gene is APETALA1(AP1), the B genes are APETALA3 (AP3) and PISTILLATA(PI) and the C gene is AGAMOUS(AG) [3–8]. Subsequent cloning of
AP1, AP3, PIand AGshow that they are all members of the MADS-box gene family and are expressed only in regions of the developing flowers that require their activities (Figure 1, reviewed in [9]). Although these results explain their region-specific requirement in specifying floral organ identities, they also raise the issue of how the floral homeotic genes are activated in floral meristems, and how they come to be expressed in spatially restricted domains.
In order to address these issues, genes acting upstream of the floral homeotic genes must be identified. The meris-tem identity genes, which include LEAFY (LFY),
UNUSUAL FLORAL ORGANS(UFO) and AP1, are excel-lent candidates for such upstream acting genes [3,7,10–14]. Plants carrying mutations in any of the meristem identity genes produce flowers with shoot-like characters, which suggests these genes normally instruct the meristems to adopt a floral fate. As the determination of floral meristem identity precedes that of floral organ identity, the meristem identity genes must act upstream of the floral homeotic genes. LFYencodes a novel transcription factor, whereas
UFOencodes an F-box-containing protein [12,14,15,16••].
LFY, UFOand AP1are expressed very early during flower development, consistent with the idea that they can acti-vate the expression of the floral homeotic genes (Figure 1). Three recent papers now convincingly show that AP1and
AGare direct targets of LFY, and that LFYactivates AP1,
AP3and AGusing different mechanisms [16••–18••].
AG
is a direct target of
LFY
Mutations in AGresult in flowers with third-whorl petals, and the fourth whorl develops as a new agmutant flower [4]. Transcripts of the C class gene AGcan be detected in the center of a wild-type flower from floral stage 3, corre-sponding to whorls three and four (Figure 1, [4,19]). The first indication that LFYmay be critical for AGexpression came from analyzing the expression pattern of AGin strong
lfy mutants, in which the early arising flowers are trans-formed into leaves with associated shoots, whereas the later arising flowers develop bracts in whorl one, lack petals and stamens, and develop irregularly fused carpels in the center [12,20,21]. In the later arising flowers the onset of AGexpression is delayed, although AGeventually accumulates in the center [22].
To begin probing into the mechanism by which LFY acti-vates AG expression, Parcy et al. [16••] generated a gain-of-function LFYallele, called LFY:VP16, that is consti-tutively active by inserting the transcriptional activation domain of VP16 into LFY. This experiment aimed to address whether the role of LFYin activating floral homeotic genes
Three ways to learn the ABCs
could be separated from its earlier role in specifying the iden-tity of floral meristems. It turned out that LFY:VP16
transgenic plants have unaltered ability to initiate flowers, whereas the flower morphology is clearly affected, implicat-ing LFYin regulating floral homeotic genes.
As with all gain-of-function alleles, it is important to show that the LFY:VP16phenotypes reveal the normal functions of the endogenous LFYgene. Therefore, Parcy et al. [16••] performed two elegant control experiments. First, the
LFY:VP16 phenotypes are enhanced by decreasing the gene dosage of the endogenous LFY, which strongly sug-gests that LFY:VP16 competes for the same target genes as the endogenous LFY. Furthermore, LFY:VP16rescues the flower initiation defects of lfymutants. Second, a mutant
LFY:VP16version (called LFY:mVP16), in which a mutant VP16 domain that is inactive in transcriptional activation was inserted into LFY, fully rescues lfy mutants but does not affect floral morphology. These results show that inser-tion of a foreign polypeptide per se does not activate or inactivate the LFY protein.
A detailed examination of LFY:VP16 plants shows that sepals are converted to carpels, and petals to stamens [16••]. In short, they resemble plants constitutively expressing AG
[23]. RNA in situhybridizations confirm that AGexpression is ectopic, precocious and at high levels in LFY:VP16plants.
This observation is consistent with the fact that ubiquitous expression of LFY:VP16in the vegetative tissue also leads to parallel ubiquitous expression of AG.
If LFY:VP16can activate AGin all tissue types, why does LFY, which is present throughout the floral meristem from floral stages 1 to 3, normally induce AGonly in the center of the flower [12,13,16••]? Parcy et al. [16••] have proposed two models to explain this conundrum. First, a repressor activity, such as APETALA2, is present in the periphery of the floral meristem and prevents LFYfrom activating AG
expression [24]. Second, the repressor activity is selective-ly overcome in the center of the stage 3 floral meristem. In either model, regional expression of AG requires LFY, which provides floral meristem-specific cue, and an unde-fined molecule ‘X’, which provides the C-region cue. Parcy
et al. [16••] have suggested further that the VP16 transcrip-tional activation domain renders the LFY protein independent of other regulators of AG, leading to the acti-vation of AGexpression throughout the flower.
In a follow-on study, LFYhas been shown to activate AG
expression directly by binding to an enhancer in the first intron of AG[17••]. It was previously demonstrated that the first intron of AGis crucial for its expression [25]; Bush et al. [17••] have gone on to show that transcriptional enhancers present in the first intron of AGare sufficient to confer a Figure 1
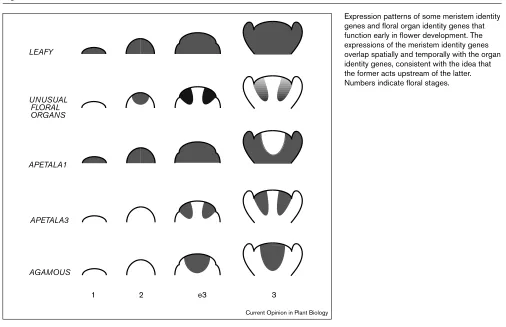
Expression patterns of some meristem identity genes and floral organ identity genes that function early in flower development. The expressions of the meristem identity genes overlap spatially and temporally with the organ identity genes, consistent with the idea that the former acts upstream of the latter. Numbers indicate floral stages. UNUSUAL
FLORAL ORGANS LEAFY
APETALA3 APETALA1
1 2 e3 3
AGAMOUS
wild-type AGexpression pattern. They embarked on a ‘tour de force’ approach to define the smallest piece of DNA that retains full activities of the AG enhancer, and found that two non-overlapping fragments from the intron can confer
AG-specific expression. Busch et al. [17••] decided to focus on the smaller 3′enhancer because its expression seems to be less complex. Further deletions of the 3′enhancer led to progressive reduction of its activity; however, LFY respon-siveness could be followed using LFY:VP16. This approach defines a LFYresponsive element to a 230 bp region. Using an in vitro DNA-binding assay, two closely spaced LFY binding sites were identified in the 230 bp fragment of the
AG intron. To assess the functional significance of these sites in vivo, a small deletion including the LFY binding sites, and point mutations abolishing LFY binding in vitro
were introduced into the AG3′enhancer. Mutating both binding sites inactivates the enhancer, whereas mutating one site significantly attenuates the enhancer. Taken together, these results show that LFY is a direct upstream activator of AGexpression.
AP1
is also a direct target of
LFY
AP1 is both a meristem identity gene and an A function organ identity gene, as mentioned above. Plants homozy-gous for strong ap1alleles develop bracts in the first floral whorl, usually lack petals, and have secondary flowers in the axils of the first floral organs [3,7]. AP1 is initially expressed throughout the floral meristem from floral stages 1 to 3 (Figure 1, [6]). Expression then abates in the two central whorls because of negative regulation by AG[6,26]. To explain how AP1is expressed in the outer two whorls of a mature flower, therefore, one needs to explain how AP1
is initially expressed throughout the floral meristem.
Numerous experiments suggest that AP1activity is mostly downstream of LFY. AP1 expression is significantly delayed and reduced in lfy mutants [27•,28•]. Constitutive expression of LFY (35S::LFY) leads to precocious expres-sion of AP1 [29]. In addition, ap1 mutants attenuate the shoot-to-flower conversion phenotype of the 35S::LFY plants, whereas the gain-of-function phenotype of 35S::AP1
plants is mostly unaffected by mutations in LFY
[27•,29,30]. These observations prompted Parcy et al. [16••] to examine AP1expression in LFY:VP16plants, although the phenotype of these plants suggests no a priory reason for altered AP1 expression. They found that the level of early AP1expression is greatly elevated, although its early pattern of expression is unaltered. In vitro DNA-binding assays showed that a high-affinity LFY binding site is pre-sent in the AP1promoter [16••,17••]. Although this site is present in a minimal AP1promoter, its in vivofunction is unknown [31]. Consistent with the idea that AP1 expres-sion is directly regulated by LFY, LFY:VP16 activates the expression of a reporter gene in yeast under the control of an AP1promoter containing the LFY binding site [16••].
Another recent study provides evidence that LFY is a direct transcriptional activator of AP1 in vivo [18••].
Wagner et al. [18••] fused the hormone-binding domain of the rat glucocorticoid receptor to LFY [32]. The chimeric protein is expressed constitutively under the control of the 35S promoter (35S::LFY-GR). In the absence of glucocor-ticoid, LFY-GR should be tethered in the cytoplasm by interaction with the chaperon proteins, rendering the tran-scription factor LFY inactive [18••,32,33•]. Upon treatment with glucocorticoid, LFY-GR dissociates from the chaperons, translocates to the nucleus, and regulates target gene expression. As activation of LFY-GR is post-translational, the immediate effect of LFY activation on transcription of any LFYtarget genes can be monitored in the presence of a protein synthesis inhibitor.
The 35S::LFY-GR construct was introduced into strong lfy
mutants. To show that the translational fusion does not compromise LFY activity, Wagner et al. [18••] examined the phenotype of these plants upon dexamethasone (a strong glucocorticoid) treatment. They found that, after such treatment, 35S::LFY-GR mostly rescues the floral morphology of lfy mutants. In addition, an early flowering phenotype and the shoot-to-flower conversions were observed in these dexamethasone treated plants; there-fore, 35S::LFY-GR has the same activities as 35S::LFY [18••,29]. To evaluate whether AP1 is a direct target of
LFY, AP1expression was analyzed in lfy mutants carrying the 35S::LFY-GR transgene treated with dexamethasone and cycloheximide. Cycloheximide treatment, which pre-vents protein synthesis, ensures that only genes directly activated by LFYare induced upon dexamethasone addi-tion. AP1 RNA could be detected in young flower primordia of these plants eight hours after dexamethasone and cycloheximide treatment.
Activation of
AP3
by
LFY
and
UFO
Plants carrying mutations in AP3or PIhave sepals in the second and carpels in the third whorl [5,8]. AP3starts to be expressed in the second and third whorls from floral stage 3 (Figure 1, [5]). Whereas the initiation of AP3expression is unchanged in ap3 and pimutants, continued expression of AP3after floral stage 6 depends on wild-type activities of AP3and PI[5,34].
Several lines of evidence suggest that LFYand UFO are key upstream regulators of AP3. Flowers from strong lfy
and ufo mutants lack petals and stamens [12–14,20,21];
AP3expression is significantly reduced in strong lfy and ufo
mutants [13,22]; and constitutive expression of AP3and PI
partially restores stamens and petals in lfy and ufo mutants, whereas constitutive expression of UFO does not rescue
ap3 and pimutants [15,35].
Consistent with its upstream regulatory role, UFO RNA accumulates in the floral meristem before the onset of AP3
of UFORNA in the center. During late floral stage 3, UFO
RNA can be detected in the second and third whorls, sim-ilar to the AP3 expression domain at the same stage. During floral stage 4, the UFO domain becomes mainly restricted to the petal primordia. From embryonic to repro-ductive phases of development, UFOis also expressed at high levels at the periphery and at low levels at the center of the SAM [15,37].
How do LFYand UFOactivate AP3 expression? Clearly, the simple hierarchical models of UFOacting downstream of LFYand LFYacting downstream of UFOare incorrect. This is because constitutive expression of UFOfails to res-cue lfy mutants, and, conversely, constitutive expression of
LFY does not rescue ufo mutants [15,29]. Plants doubly transgenic for 35S::LFY and 35S::UFO have ubiquitous expression of AP3throughout the developmentally arrest-ed searrest-edings [16••]. In contrast, AP3cannot be detected in seedlings expressing either LFYor UFO. On the basis of these results, Parcy et al. [16••] have suggested that during normal development the expression domain of AP3 is defined by LFY, which is expressed throughout the devel-oping flower, and UFO, which is expressed in the emerging petal and stamen primordia. In this context, LFYprovides the floral meristem specificity and UFO provides the regional specificity for AP3 expression, analogous to the proposal that LFYand the unknown factor ‘X’ activate AG
expression in the center of the flower.
Although this model provides a good framework for AP3
activation, some results cannot be easily reconciled with it. Much evidence has shown that co-expression of LFYand
UFOis not sufficient for AP3expression. First, the shoot apex of 35S::LFY plants, which expresses endogenous
UFO, does not express AP3[16••]; similarly, AP3cannot be detected in the shoot apex of plants expressing LFYunder the control of UFO promoter [15]. Second, constitutive expression of UFOfails to initiate AP3expression during floral stage 1 of the floral primordia, which express high levels of LFY[15]. These observations led Lee et al. [15] to propose that induction of AP3expression by UFOand
LFY is dependent on additional factors. It should be emphasized that the model proposed by Parcy et al. [16••] is not necessarily rejected by the results described above. For example, high levels of both LFYand UFOmay be required for AP3 expression, and this condition may be achieved only in seedlings expressing LFYand UFOunder the control of the 35S promoter, and in wild-type flowers.
At present, it is unclear whether LFYdirectly activates AP3
transcription. However, it should be very straightforward to address this issue by analyzing the lfy 35S::LFY-GR plants. If these plants carry an AP3::GUSreporter gene, they will stain positive for GUS activity after dexamethasone treatment [18••], and, if AP3is a direct target of LFY, then AP3and
GUSRNA should be detected in these plants after treatment with dexamethasone and cycloheximide. Regulatory ele-ments crucial for AP3 expression are beginning to be
identified, therefore, one might also start looking for LFY-binding sites in the AP3promoter [38•,39•].
As UFO is not a transcription factor, it is unlikely to direct-ly activate AP3transcription. Analyses of the Antirrhinum UFOorthologue FIMBRIATA(FIM) provide good insights into how UFOmay function [40]. Antirrhinumproteins with strong similarity to Skp1 from yeast and animals have been shown to interact with FIM [40]. In yeast and humans, Skp1 proteins interact with F-box containing proteins to form a complex targeted for degradation, which is required for cell cycle progression [41–43]. Furthermore, it has also been shown that an F-box protein, E3RSIκB, targets IκB, a repressor of the transcription factor NK-κB, for ubiquitin-proteasome-mediated degradation [44]. On the basis of these results, it is tempting to speculate that a protein com-plex formed by UFO and the Arabidopsis Skp1-like proteins might act by promoting degradation of a tran-scriptional repressor of AP3. The absence of the repressor in the second and third whorls, and the presence of the activator LFY throughout the floral meristem may be nec-essary for spatially restricted AP3expression.
Conclusions
While significant progress has been made toward elucidat-ing how the floral homeotic genes are expressed in spatially restricted patterns, our understanding of the transcriptional regulation of these genes is clearly incomplete. We now know that the LFY meristem identity gene directly acti-vates AP1and AG, and perhaps AP3, although these studies indicate that additional factors (e.g.,UFO, factor ‘X’) must also interact with LFY in this process. We also know that many other genes are involved in regulating the ABC genes, including AP1, AP2, CAULIFLOWER, CURLY LEAF, SUPERMANand LEUNIG, although their mecha-nistic roles have yet to be clearly defined [7,22,45–50]. Given the long-term goal of elucidating the cascade of tar-get gene regulation beginning in the floral meristem with genes such as LFY, and ending with fully differentiated flo-ral organs, it is clear that we are only scratching the surface of what promises to be a very deep and interesting story.
Acknowledgements
We thank François Parcy and Detlef Weigel for comments. MN received a long-term post-doctorate fellowship from The Human Frontier Science Program Organization (LT-367/97). Research in the laboratory of MFY is supported by grants from the National Science Foundation and the National Institutes of Health.
References and recommended reading
Papers of particular interest, published within the annual period of review, have been highlighted as:
• of special interest
••of outstanding interest
1. Coen ES, Meyerowitz EM: The war of the whorls: genetic interactions controlling flower development.Nature1991, 353:31-37.
2. Weigel D, Meyerowitz E: The ABCs of floral homeotic genes.Cell
1994, 78:203-209.
4. Yanofsky MF, Ma H, Bowman JL, Drews GN, Feldmann KA, Meyerowitz EM: The protein encoded by the Arabidopsishomeotic gene agamousresembles transcription factors.Nature1990, 346:35-39.
5. Jack T, Brockman LL, Meyerowitz EM: The homeotic gene APETALA3of Arabidopsis thalianaencodes a MADS box and is expressed in petals and stamens.Cell1992, 68:683-697.
6. Mandel MA, Gustafson-Brown C, Savidge B, Yanofsky MF: Molecular characterization of the Arabidopsisfloral homeotic gene APETALA1.Nature1992, 360:273-277.
7. Bowman JL, Alvarez J, Weigel D, Meyerowitz EM, Smyth DR: Control of flower development in Arabidopsis thalianaby APETALA1and interacting genes.Development1993, 119:721-743.
8. Goto K, Meyerowitz EM: Function and regulation of the Arabidopsis floral homeotic gene PISTILLATA.Genes Dev1993, 8:1548-1560.
9. Riechmann JL, Meyerowitz EM: MADS domain proteins in plant development.Biol Chem1997, 378:1079-1101.
10. Schultz EA, Haughn GW: LEAFY, a homeotic gene that regulates inflorescence development in Arabidopsis.Plant Cell1991,
3:771-781.
11. Huala E, Sussex IM: LEAFYinteracts with floral homeotic genes to regulate Arabidopsisfloral development.Plant Cell1992, 4:901-913.
12. Weigel D, Alvarez J, Smyth DR, Yanofsky MF, Meyerowitz EM: LEAFY controls floral meristem identity in Arabidopsis.Cell1992,
69:843-859.
13. Levin JZ, Meyerowitz EM: UFO: an Arabidopsis gene involved in both floral meristem and floral organ development.Plant Cell
1995, 7:529-548.
14. Wilkinson MD, Haughn GW: UNUSUAL FLORAL ORGANScontrols meristem identity and organ primordia fate in Arabidopsis.
Plant Cell1995, 7:1485-1499.
15. Lee I, Wolfe DS, Nilsson O, Weigel D: A LEAFYco-regulator encoded by UNUSUAL FLORAL ORGANS.Curr Biol1997, 7:95-104.
16. Parcy F, Nilsson O, Busch MA, Lee I, Weigel D: A genetic framework
•• for floral patterning.Nature1998, 395:561-566.
This paper describes the phenotype of plants carrying a gain-of-function allele of LEAFY, called LFY:VP16, which was constructed by inserting the transcriptional activation domain of VP16 into LFY. It was found that
LFY:VP16has different effects on the expression of the A, B and C class flo-ral homeotic genes. The authors conclude that LFY activates different floflo-ral homeotic genes using different mechanisms.
17. Busch MA, Bomblies K, Weigel D: Activation of a floral homeotic
•• gene in Arabidopsis.Science1999, 285:585-587.
This is an extension of the previous study [16••]. The authors analyze the
enhancer elements controlling AGAMOUSexpression. They find that two
LEAFYbinding sites in the first intron of AGare necessary for LFYdirected
AGexpression in vivo. Therefore, LFYis formally a direct upstream activator of AGexpression.
18. Wagner D, Sablowski RWM, Meyerowitz EM: Transcriptional
•• activation of APETALA1 by LEAFY.Science1999, 285:582-584. By using a steroid-inducible LFY, called 35S::LFY-GR, the authors show that early, but not late, expression of APETALA1results from direct transcrip-tional activation by LEAFY.
19. Drews GN, Bowman JL, Meyerowitz EM: Negative regulation of the Arabidopsishomeotic gene AGAMOUSby the APETALA2product.
Cell1991, 65:991-1002.
20. Schultz EA, Haughn GW: LEAFY, a homeotic gene that regulates inflorescence development in Arabidopsis.Plant Cell1991,
3:771-781.
21. Haula E, Sussex IM: LEAFYinteracts with floral homeotic genes to regulate Arabidopsis floral development.Plant Cell1992,
4:901-913.
22. Weigel D, Meyerowitz EM: Activation of floral homeotic genes in Arabidopsis.Science1993, 261:1723-1726.
23. Mizukami Y, Ma H: Ectopic expression of the floral homeotic gene AGAMOUSin transgenic Arabidopsisplants alters floral organ identity.Cell1992, 71:119-131.
24. Bowman JL, Smyth DR, Meyerowitz EM: Genetic interactions among floral homeotic genes of Arabidopsis.Development1991,
112:1-20.
25. Sieburth LE, Meyerowitz EM: Molecular dissection of the AGAMOUS control region shows that cis elements for spatial regulation are located intragenically.Plant Cell1997, 9:355-365.
26. Gustafson-Brown C, Savidge B, Yanofsky MF: Regulation of the Arabidopsisfloral homeotic gene APETALA1.Cell1994, 76:131-143.
27. Liljegren SJ, Gustafson-Brown C, Pinyopich A, Ditta GS,
• Yanofsky MF: Interactions among APETALA1, LEAFY, and TERMINAL FLOWER1specify meristem fate.Plant Cell1999,
11:1007-1018.
Interactions among APETALA1, LEAFYand TERMINAL FLOWER1are described in this paper. The authors show that AP1can activate LFY expres-sion, and vice versa. By contrast, AP1and TFL1negatively regulate each other’s expression. It is concluded that these interactions contribute to the sharp transition that occurs from vegetative to reproductive growth phases. 28. Ratcliffe OJ, Bradley DJ, Coen ES: Separation of shoot and floral
• identity in Arabidopsis.Development1999, 126:1109-1120. This paper describes the hierarchy among several genes (APETALA1, CAU-LIFLOWER, LEAFYand TERMINAL FLOWER 1) which affect the shoot meristems and floral meristems identities. The meristem identity genes (AP1,
CALand LFY) prevent TFL1transcription in the floral meristems; converse-ly, TFL1delays the upregulation of the meristem identity genes, and prevents the shoot meristems from responding to LFYand AP1. The authors con-clude that the relative timing of upregulating TFL1and the meristem identity genes determine their final expression patterns.
29. Weigel D, Nilsson O: A developmental switch sufficient for flower initiation in diverse plants.Nature1995, 377:495-500.
30. Mandel MA, Yanofsky MF: A gene triggering flower formation in Arabidopsis.Nature1995, 377:522-524.
31. Hempel FD, Weigel D, Mandel MA, Ditta G, Zambryski PC, Feldman L, Yanofsky MF: Floral determination and expression of floral regulatory genes in Arabidopsis.Development1997,
124:3845-3853.
32. Lloyd AM, Schena M, Walbot V, Davis RW: Epidermal cell fate determination in Arabidopsis: patterns defined by a steroid-inducible regulator.Science1994, 266:436-439.
33. Sablowski RWM, Meyerowitz EM: A homolog of NO APICAL
• MERISTEMis an immediate target of the floral homeotic genes APETALA3/PISTILLATA.Cell1998, 92:93-103.
APETALA3and PISTILLATAfunction are placed under post-translational control by use of a steroid-inducible AP3 (35S::AP3-GR). Using differential display, three direct targets of AP3/PI are identified and one of them is called NAP(NO APICAL MERISTEM-LIKE, ACTIVATED BY AP3/PI). The expression pattern of NAPand the phenotypes caused by its mis-expression suggest that NAPis important for cell division and cell expansion in stamens and petals.
34. Jack T, Fox GL, Meyerowitz EM: Arabidopsishomeotic gene APETALA3ectopic expression: transcriptional and
posttranscriptional regulation determine floral organ identity.Cell
1994, 76:703-716.
35. Krizek BA, Meyerowitz EM: The Arabidopsis homeotic genes APETALA3and PISTILLATAare sufficient to provide the B class organ identity function.Development1996, 122:11-22.
36. Ingram GC, Goodrich J, Wilkinson MD, Simon R, Haughn GW, Coen ES: Parallels between UNUSUAL FLORAL ORGANSand FIMBRIATA, genes controlling flower development in Arabidopsis and Antirrhinum.Plant Cell1995, 7:1501-1510.
37. Long JA, Barton MK: The development of apical embryonic pattern in Arabidopsis.Development1998, 125:3027-3035.
38. Hill TA, Day CD, Zondlo SC, Thackeray AG, Irish VF: Discrete spatial
• and temporal cis-acting elements regulate transcription of the Arabidopsisfloral homeotic gene APETALA3.Development1998,
125:1711-1721.
This paper describes molecular dissection of the APETALA3promoter. The authors identify regions of the promoter require for petal-specific and sta-men-specific expression, and show that AP3, PISTILLATA, UNUSUAL FLO-RAL ORGANSand APETALA1are required for AP3::GUSexpression in the petals. The authors also demonstrate that AP1, AP3, PI and AG bind to three sequence elements, called CArG boxes, present in the AP3promoter. See also [39•].
39. Tilly JJ, Allen DW, Jack T: The CArG boxes in the promoter of the
• Arabidopsisfloral organ identity gene APETALA3mediate diverse regulatory effects.Development1998, 125:1647-1657.
To understand how the expression of APETALA3is regulated, the authors ana-lyze the AP3promoter using AP3::GUSfusions. A 496 bp fragment of the AP3
143 bp fragment, directs GUS activity in the same spatial and temporal expres-sion pattern as the endogenous AP3gene. Mutations of the three CArG (CArG1–CArG3) boxes, which are present in the 143 bp fragment, affects GUS expression in the context of the synthetic promoter. The authors conclude that CArG1 binds a positively acting factor(s), CArG2 is required for petal spe-cific expression, and CArG3 binds a negatively acting factor(s). See also [38•].
40. Ingram GC, Doyle S, Carpenter R, Schultz EA, Simon R, Coen ES:
Dual role for fimbriatain regulating floral homeotic genes and cell division in Antirrhinum.EMBO J1997, 16:6521-6534.
41. Zhang H, Kobayashi R, Galaktionov K, Beach D: p19Skp1and p45Skp2
are essential elements of the cyclinA-CDK2 S phase kinase.Cell
1995, 82:915-925.
42. Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper JW, Elledge S: SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box.Cell1996, 86:263-274.
43. Connelly C, Hieter P: Budding yeast SKP1encodes an evolutionarily conserved kinetochore protein required for cell cycle progression.Cell1996, 86:275-285.
44. Yaron A, Hatzubai A, Davis M, Lavon I, Amit S, Manning AM, Andersen JS, Mann M, Mercurio F, Ben-Neriah Y: Identification of the
receptor component of the IkBa-ubiquitin ligase.Nature1998,
396:590-594.
45. Bowman JL, Sakai H, Jack T, Weigel D, Mayer U, Meyerowitz EM:
SUPERMAN, a regulator of floral homeotic genes in Arabidopsis.
Development1992, 114:599-615.
46. Jofuku DK, den Boer BGW, Van Montagu M, Okamuro JK: Control of Arabidopsisflower and seed development by the homeotic gene APETALA2.Plant Cell1994, 6:1211-1225.
47. Kempin SA, Savidge B, Yanofsky MF: Molecular basis of the cauliflowerphenotype in Arabidopsis.Science1995, 267:522-525.
48. Liu Z, Meyerowitz EM: LEUNIGregulates AGAMOUSexpression in Arabidopsis.Development1995, 121:975-991.
49. Sakai H, Medrano LJ, Meyerowitz EM: Role of SUPERMANin maintaining Arabidopsis floral whorl boundaries.Nature1995,
378:199-203.