PDBe Protein Data Bank in Europe (1)
Bebas
8
0
0
Teks penuh
Gambar
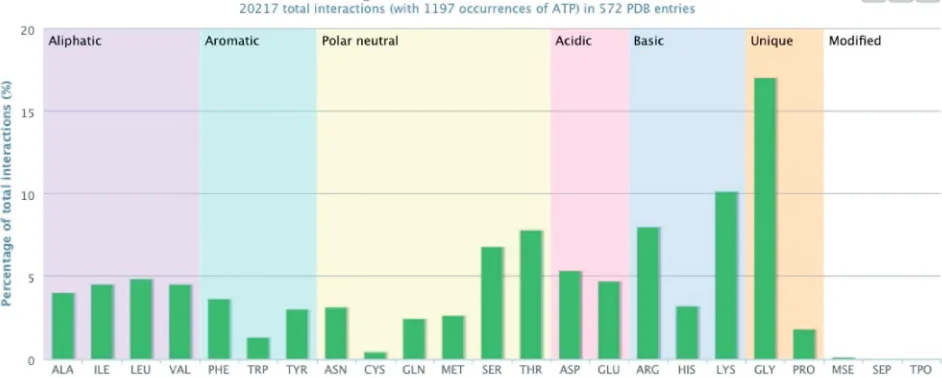
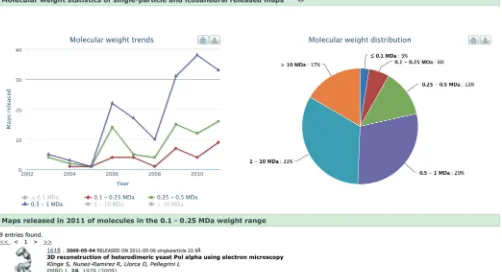
+3
Dokumen terkait