Molecular cloning and characterization of a plant homologue of
the origin recognition complex 1 (ORC1)
Seisuke Kimura
a, Toyotaka Ishibashi
a, Masami Hatanaka
a, Yoshikiyo Sakakibara
b,
Junji Hashimoto
c,d, Kengo Sakaguchi
a,*
aDepartment of Applied Biological Science,Faculty of Science and Technology,Science Uni6ersity of Tokyo,2641Yamazaki,Noda-shi, Chiba-ken278-8510,Japan
bMolecular Function Laboratory,National Food Research Institute,Tsukuba-shi,Ibaraki-ken305-8642, Japan cNational Institute of Agrobiological Resources,Tsukuba-shi,Ibaraki-ken305-8642,Japan
dCREST (Core Research for E6olutional Science and Technology)of Japan Science and Technology Corporation(JST),Tsukuba-shi, Ibaraki-ken305-8642,Japan
Received 8 March 2000; accepted 22 May 2000
Abstract
By using the rice EST database, we have isolated a 2.8 kb cDNA, termedOryza sati6a ORC1 (OsORC1), from rice (O.sati6a)
encoding a protein that shows homology with the eukaryotic ORC1 proteins. Alignment of theOsORC1 protein sequence with the sequence of ORC1 from human and yeastsS.cere6isiaeandS.pombeshowed a high degree of sequence homology (38.7, 32.9
and 35.0% identity, respectively), particularly around the C-terminal region containing the CDC-NTP domain. Interestingly, the
OsORC1 protein had an A+T hook-like motif, which was not present in the human or yeast genes. Genomic analysis indicated thatOsORC1 existed as a single copy per genome.OsORC1 transcripts were expressed strongly in root tips and weakly in young leaves containing root apical meristem and marginal meristem, respectively. No expression was detected in the mature leaves. The level ofOsORC1 expression was significantly reduced when cell proliferation was temporarily halted by the removal of sucrose from the growth medium. When the growth-halted cells began to re-grow following addition of sucrose to the medium,OsORC1 was again expressed at high levels. These results suggested thatOsORC1 is required for cell proliferation. The role ofOsORC1 in plant DNA replication will be discussed. © 2000 Elsevier Science Ireland Ltd. All rights reserved.
Keywords:Oryza sati6a; Origin recognition complex; A+T hook-like motif; DNA replication and repair
www.elsevier.com/locate/plantsci
1. Introduction
DNA replication and repair are important pro-cesses for the maintenance of genomic integrity, and the coordination between cell cycle-regulated DNA replication and DNA repair is required to avoid mutation and maintain genomic integrity.
Our research interest is in the mechanism of coor-dination of plant DNA replication and repair [1 – 4].
Higher plants are an interesting system in which to study the mechanisms of DNA replication and their relationship to DNA repair. The normal development of higher plants is regulated by the initiation of cell proliferation of the meristematic tissue, and organ formation in plants is accom-plished by a combination of oriented cell divisions and tissue formation. Plants are exposed to severe UV for much longer periods than animals or yeast [1]. The meristematic tissue and the organs on the
* Corresponding author. Tel.: +81-471-241501 ext. 3409; fax:
+81-471-239767.
E-mail address:[email protected] (K. Sakaguchi).
ground must proliferate under UV bombardment more severe than that to which animals or yeast are exposed. Stapleton et al. [5] measured cyclobu-tane pyrimidine dimers in maize DNA from plants grown under natural solar radiation. Green maize tissues (leaf sheath and leaf blade) showed de-tectable DNA damage, roots were less damaged, and anthers were much more damaged than the green leaves [5]. The UV-damaged DNA must be repaired rapidly at the sites at which DNA replica-tion occurs. Thus, higher plants might be a good system in which to investigate the relationship between DNA replication and DNA repair, espe-cially in response to UV bombardment. There is little information about the mechanism of coordi-nation of DNA replication and the repair in plant. Also, little is known about the proteins or genes involved in DNA replication and repair in higher plants in comparison with animals or lower eu-karyotes [1 – 4,6 – 10]. From this point of view, we reported the molecular cloning and characteriza-tion of FEN-1 in the higher plant, rice (Oryza sati6a L. Nipponbare) [1].
Understanding the mechanism of coordination of DNA replication and repair requires the deter-mination of the mechanism underlying regulation of initiation of DNA replication. The initiation of DNA replication is a dynamic process that occurs through the coordinated assembly and disassembly of protein complexes at the replication origins [11 – 15]. Originally identified as a multi-subunit complex that specifically binds to the DNA repli-cation origins in S. cere6isiase [16,17], the origin
recognition complex (ORC) is a critical factor required for the initiation of eukaryotic DNA replication. ORC is bound to the origins of chro-mosomal DNA replication throughout the cell cycle and is thought to function as a binding site for a series of cell cycle regulated protein com-plexes [11 – 15,18]. In addition to its role in DNA replication, ORC also functions in transcriptional silencing at the yeast silent mating type loci HML and HMR [19]. Sequence analogs of ORC sub-units have been identified in various organisms suggesting that the ORC complex has been con-served during evolution [20 – 24]. Studies of the plant counterpart could provide valuable insight into the mechanism of coordination of DNA repli-cation and repair. In this study, we isolated and characterized a plant homologue of ORC1.
2. Materials and methods
2.1. Plant material
Rice plants (O. sati6a L. cv. Nipponbare) were grown in a growth cabinet under a 16 h day/8 h night cycle at 28°C. Rice cells were grown in suspension culture as described earlier [25].
2.2. Southern hybridization
Rice genomic DNA was digested with BglII,
EcoRI, HindIII or XbaI, and the DNA digests were resolved on 1% agarose gels. The restriction fragments were transferred onto nylon membrane (Hybond-N, Amersham) according to the manu-facturer’s recommendations. A 32P-labeled probe was prepared by random priming of O. sati6a
ORC1 (OsORC1) cDNA (Rediprime II,
Amer-sham). After prehybridization, hybridization was carried out at 42°C for 16 h, followed by washing twice with 6×SSC +1% SDS at room tempera-ture for 15 min, twice with 1×SSC+1% SDS at 65°C for 15 min, and finally twice with 0.1×
SSC+1% SDS at 65°C for 15 min.
2.3. Northern hybridization
Aliquots of 20 mg of total RNA were resolved on 1.2% formaldehyde agarose gels and trans-ferred onto nylon membranes (Hybond-N, Amer-sham). After prehybridization, the filters were probed with 32P-labeled OsORC1 cDNA at 42°C for 16 h and followed by washing twice with 2×SSC+0.1% SDS at room temperature for 15 min, and three times with 0.1×SSC+0.1% SDS at 65°C for 20 min.
2.4. Analysis of DNA sequences
3. Results and discussion
3.1. Molecular cloning of Oryza sati6a ORC1 homologue using the EST database
The rice EST database was searched using the tBLASTn program to identify cDNA clones with homology to the origin recognition complex 1 (ORC1) protein. Rice EST clone S3529 (GenBank accession no. D41200) was found to have signifi-cant homology. The insert DNA of the EST clone was excised and used to screen the rice cDNA library by the plaque hybridization method. Many independent plaques produced strong hybridiza-tion signals. The insert DNAs were excised as pBluescript SK+ phagemids and sequenced. Screening of a rice cDNA library with the frag-ment as a probe resulted in isolation of a 2.8-kb clone designated as OsORC1 (O. sati6a ORC1).
The clone possessed one potential translational initiation site (ATG; nucleotides 102 – 104), one in-frame termination codon (TAA; nucleotides 2440 – 2442), and a 3%poly (A) tail. The open read-ing frame of OsORC1 encoded a predicted product of 779 amino acid residues with a molecu-lar mass of 88.1 kDa (Fig. 1), simimolecu-lar in size to other eukaryotic ORC1 homologues, and with an estimated pI of 8.25. The nucleotide sequence data reported in this paper appear in the DDBJ/
EMBL/GenBank nucleotide sequence databases with the accession number AB037135.
3.2. Comparison of OsORC1 to other eukaryotic ORC1 proteins
The deduced amino acid sequence of OsORC1 was compared with the sequences of the human [20], Xenopus [22], C. albicans (Accession No.
Fig. 2. (A) Presence of an A+T-hook-like motif in the N-terminal region ofOsORC1. (B) Multiple alignment of ORC1 proteins across the CDC-NTP domain. S. p., S. c., K. l. and C. albicans indicate S. pombe, S. cere6isiae, K. lactis and C. alibicans,
respectively.
O74270), K. lactis [20], S. pombe [20] and S.
cere6isiae [23] homologues (Figs. 1 and 2). The
sequence showed sequence identity with human (38.7%),Xenopus(34.1%),S.cere6isiae(32.9%), and
S. pombe (35.0%) ORC1 homologues.
The OsORC1 protein shared high degrees of conservation in amino acid sequences with the human ORC1 in the side of the C-terminal domain from amino acids 435 to 670 (Figs. 1 – 3), which is separated into six smaller boxes, Box 1 – 6, known as the cell division cycle-nucleoside triphosphate-binding protein (NTP) domain. The CDC-NTP domain is required for DNA binding activity to replication origins, and showed a high degree of homology with those of mammals and yeasts (Figs. 1 and 2). Interestingly, the N-terminal region of the deduced amino acid sequence contained a copy of
an A+T hook-like motif (KRQRQRQRQQ;
amino acids 106 – 115) that was not present in human and yeasts (Figs. 1 and 2). According to Chuang et al. (Abstracts at the 1999 Eukaryotic DNA Replication Meeting in Cold Spring Harbor Symposium), inORCcomplex of yeasts one of the
The deduced amino acid sequence of OsORC1 was compared with those of other known eukary-otic ORC1 proteins to determine their phyloge-netic relationships and the unrooted tree was drawn based on the alignment. The two vertebrate enzymes, human ORC1 and Xenopus ORC1, are closely related to each other, and S. cere6isiae
ORC1,S.pombeORC1,C. albicansORC1 andK.
Fig. 5. Expression ofOsORC1 in different organs. Each lane contained 20 mg of total RNA isolated from mature leaves
(lane 1), young leaves (lane 2), roots (lane 3) or root tips (lane 4) from 50-day-old rice plants. The blot was probed with
32P-labeled OsORC1 cDNA (upper panel). Similar amounts
of RNA were loaded in each lane as confirmed by ethidium bromide staining (lower panel). Numbers on the right indicate the approximate length of the mRNA.
Fig. 3. Phylogenetic relationships between OsORC1 and other ORC1 proteins. Unrooted phylogenetic tree depicting the relationship among seven eukaryotic ORC1 homologues. S. c.,S.cere6isiae; S. p.,S.pombe.
lactisORC1 are distantly related to the vertebrates enzymes. OsORC1 was situated between the verte-brate and the yeast enzymes.
3.3. Genomic analysis
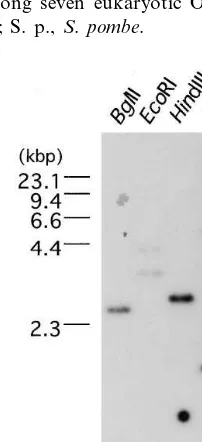
To determine the copy number of the OsORC1 gene in the genome, Southern hybridization was performed. Digested genomic DNA was hy-bridized with the full-length OsORC1 cDNA un-der high stringency conditions. As shown in Fig. 4, a single band was detected in BglII, HindIII and
XbaI digests of rice genomic DNA (lanes 1, 3 and 4 in Fig. 5), but two bands were observed in the
EcoRI digest (lane 2 in Fig. 5). These bands may have been produced by one more cuts in the introns. These results suggested that OsORC1 ex-ists as a single copy per genome.
3.4. Expression of OsORC1 in different organs of the rice plant
Next, to determine the expression pattern of
OsORC1 in various organs, northern hybridiza-tion analysis was performed. Total RNA samples isolated from various organs of 50-day-old rice plants were blotted and probed with 32P-labeled
OsORC1 cDNA. A 2.8-kb transcript was detected in the roots, especially in the root tip region. The mature and young leaves did not express the mRNA at detectable levels (lanes 1 and 2 in Fig. 5), although the young leaves have marginal meristem to increase the leaf width. Thus, Os
-Fig. 4. Southern analysis of genomic DNA from O. sati6a.
32P-labeledOsORC1 cDNA was used to probe genomic DNA
ORC1 might be expressed in the marginal meris-tem at very low levels. Since the negative signal in mature leaves was reproducible, the OsORC1 protein might not be produced in the mature leaves. The expression level in the root tips was higher than that in the whole roots (lanes 3 and 4 in Fig. 5), suggesting that OsORC1 is expressed only in root apex regions that contain root apical meristem. Cell proliferation is very active in the root meristematic tissues, and consequently the level of DNA replication in the meristem must be high. Transcription of OsORC1 might be related to the level of DNA replication.
3.5. Effects of sucrose-star6ation on the le6el of OsORC1 expression
OsORC1 was actively transcribed in rice cells in suspension culture (lane 1 in Fig. 6). When the cell proliferation was temporarily halted for 6 or 10 days by the removal of sucrose from the growth medium, the level of OsORC1 expression was sig-nificantly reduced, but not completely inhibited (lanes 2 and 3 in Fig. 6). When the growth-halted cells began to re-grow following addition of
su-crose to the medium, OsORC1 was again ex-pressed at high levels (lane 4 in Fig. 6). These results indicated that OsORC1 expression is re-quired for cell proliferation.
In this paper, we reported the molecular cloning and characterization of a plant homologue of ORC1. The origin recognition complex was origi-nally identified in the yeast S. cere6isiae as a
protein that specifically binds to origins of DNA replication. Subsequent studies suggested that ORC has important roles in several processes in-cluding DNA replication, silencing and check point control. Further studies of the biological functions of OsORC1 and other factors will yield insight into mechanism of coordination of plant DNA replication and repair.
Acknowledgements
We thank the Rice Genome Research Program (RGP) of Japan for providing the rice EST clone S3529. We also thank T. Furukawa for helpful discussions.
References
[1] S. Kimura, T. Ueda, M. Hatanaka, M. Takenouchi, J. Hashimoto, K. Sakaguchi, Plant homologue of flap en-donuclease-1: molecular cloning, characterization, and evidence for expression in meristematic tissues, Plant Mol. Biol. (2000), in press.
[2] S. Kimura, M. Takanouchi, M. Hatanaka, H. Seto, Y. Kouroku, K. Sakaguchi, An ATP-inhibited endonucle-ase from cauliflower (Brassica oleracea var. botrytis) inflorescence: purification and characterization, Planta 206 (1998) 641 – 648.
[3] H. Seto, M Hatanaka, S. Kimura, M. Oshige, Y. Tsuya, Y. Mizushina, T. Sawado, N. Aoyagi, T. Matsumoto, J. Hashimoto, K. Sakaguchi, Purification and characteriza-tion of a 100 kDa DNA polymerase from cauliflower inflorescence, Biochem. J. 332 (1998) 263 – 557.
[4] S. Kimura, M. Kai, H. Kobayashi, A. Suzuki, H. Morioka, E. Otsuka, K. Sakaguchi, A structure-specific endonuclease from cauliflower (Brassica oleracea var.
botrytis) inflorescence, Nucl. Acids Res. 28 (1997) 4970 – 4976.
[5] A.E. Stapleton, C.S. Thornber, V. Walbot, UV-B com-ponent of sunlight causes measurable damage in field grown maize (Zea maize L.): developmental cellular heterogeneity of damage and repair, Plant Cell Environ. 20 (1997) 279 – 290.
[6] A.B. Britt, Molecular genetics of DNA repair in higher plants, Trends Plant Sci. 4 (1999) 20 – 25.
Fig. 6. Effect of sucrose-starvation on the level ofOsORC1 expression. Rice cells were cultured in suspension for 6 (lanes 1 and 2) or 10 days (lane 3) with (lane 1) or without (lanes 2 and 3) sucrose, or cultured for 6 days without sucrose, then sucrose was added to the medium and culture was continued for a further 4 days (lane 4). Aliquots of 20mg of total RNA
isolated from the cultured cells were separated on 1.2% agarose gel containing formaldehyde and then blotted. The blot was probed with 32P-labeled OsORC1 cDNA. Similar
[7] M. Yokoi, M. Ito, M. Izumi, H. Miyazawa, H. Nakai, F. Hanaoka, Molecular cloning of the cDNA for the catalytic subunit of plant DNA polymerase a and its
cell-cycle dependent expression, Genes Cells 2 (1997) 695 – 709.
[8] D.T. Riberio, C.R. Machado, R.M.A. Costa, U.M. Praekelt, M.-A. Van Sluys, C.F.M. Menck, Cloning of a cDNA from Arabidopsis thaliana homologous to the humanXPBgenes, Genes 208 (1998) 207 – 213. [9] K.M. Culligan, J.B. Hays, DNA mismatch repair in
plants, Plant Physiol. 115 (1997) 833 – 839.
[10] J. Ade, F. Belzile, H. Philippe, M.P. Doutriaux, Four mismatch repair paralogues coexist in Arabidopsis thaliana: AtMSH2,AtMSH3, 1 and AtMSH6-2, Mol. Gen. Genet. 262 (1999) 239 – 249.
[11] O.M. Aparicio, D.M. Weinstein, S.P. Bell, Components and dynamics of DNA replication complexes inS.cere
-6isiae: redistribution of MCM proteins and Cdc45p during S phase, Cell 91 (1997) 59 – 69.
[12] S. Donovan, J. Harwood, L.S. Drury, J.F. Diffley, Cdc6p-dependent loading of Mcm proteins onto pre-replicative chromatin in budding yeast, Proc. Natl. Acad. Sci. 94 (1997) 5611 – 5616.
[13] C. Liang, B. Stillman, Persistent initiation of DNA replication and chromatin-bound MCM proteins during the cell cycle in cdc6 mutants, Gens. Develop. 11 (1997) 3375 – 3386.
[14] T. Tanaka, D. Knapp, K. Nasmyth, Loading of an Mcm proteins onto DNA replication origins is regulated by Cdc6p and CDKs, Cell 90 (1997) 649 – 660.
[15] J. Leatherwood, Emerging mechanisms of eukaryotic DNA replication initiation, Curr. Opin. Cell Biol. 10 (1998) 742 – 748.
[16] S.P. Bell, B. Stillman, ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex, Nature 357 (1992) 128 – 134.
[17] S.P. Bell, R. Kobayashi, B. Stillman, The multidomain structure of Orc1p reveals similarity to regulators of DNA replication and transriptional silencing, Cell 83 (1995) 1844 – 1849.
[18] J.F. Diffley, J.H. Cocker, S.J. Dowell, A. Rowley, Two steps in the assembly of complexes at the yeast replica-tion origins in vivo, Cell 78 (1964) 303 – 316.
[19] P. Laurenson, J. Rine, Silencers, silencing, and heritable transcriptional states, Microbiol. Rev. 56 (1992) 543 – 560.
[20] K.A. Gavin, M. Hidaka, B. Stillman, Conserved initia-tor proteins in eukaryotes, Science 270 (1995) 1667 – 1671.
[21] M. Muzi-Falconi, T.J. Kelly, Orp1p, a member of the cdc18/CDC6 family of S phase regulators, is ho-mologous to a component of the origin recognition complex, Proc. Natl. Acad. Sci. 92 (1995) 12475 – 12479. [22] A. Rowles, J.P. Chong, L. Brown, M. Howell, G.I. Evan, J.J. Blow, Interaction between the origin recogni-tion complex and the replicarecogni-tion licensing system in Xenopus, Cell 87 (1996) 287 – 296.
[23] S.P. Bell, R. Kobayashi, B. Stillman, Yeast origin recog-nition complex functions in transcription silencing and DNA replication, Science 262 (1993) 1844 – 1849. [24] J. Leatherwood, A. LopezGirona, P. Russel, Interaction
of cdc2 and cdc 18 with a fission yeast ORC2-like protein, Nature 379 (1996) 360 – 363.
[25] A. Baba, S. Hasegawa, K. Syono, Cultivation of rice protoplasts and their transformation mediated by
Agrobacterium spheroplasts, Plant Cell Physiol. 27 (1986) 463 – 471.