Legends for Figures S1-S8:
FIGURE S1. The seven distinct rotational isomers of DP (of a 30o torsional search) viewed along P-P projection axis.
FIGURE S2. The six conformational groups, each being represented by its extended (180o, 180o) conformation, of TP, as viewed through two adjacent P-P axes, separately.
FIGURE S3. The (t1, t1’) conformational energy surface of DP (HF/6-31G* results).
FIGURE S4. Structures of the four energy minima of MDP identified on the HF/6-31G* surface, with the shortest C-H...O distance indicated. Partial charges derived from Mulliken population analysis for each atom are labeled.
FIGURE S5. Structure of the stg-stg extended conformation of TP, the energy minimum identified on the HF/6-31G* surface.
FIGURE S6. The lowest vibrational mode and frequency associated with P-O-P bending for DP, MDP (gg I), and TP (symmetric and asymmetric), respectively. Parenthesized numbers denote the order of the particular mode in the 3N-6 vibrational modes of the molecule (3 being the third lowest, for example).
FIGURE S7. Atom labeling of the three model molecules to which the geometric parameters in Table S1 refer. Partial charges as determined by Mulliken population are those for the lowest-energy structure of each compound.
FIGURE S8. Normal mode vectors and frequencies (in cm-1) listed in increasing order for the minimum-energy structures of DP, MDP, and TP optimized with HF/6-31G*. These were created with the Gaussian94 output files and using the 啑vibs.c’ program retrieved from an archived file of the Computational Chemistry List (http://ccl.osc.edu/chemistry/html).
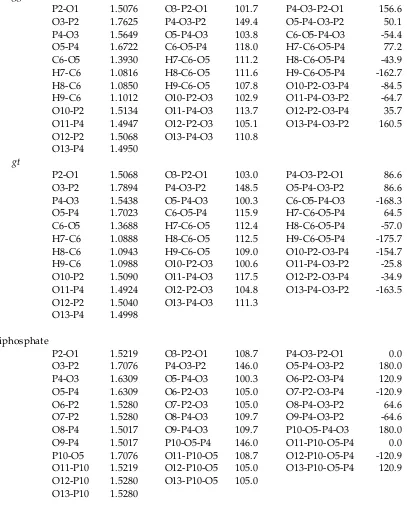
TABLE S1: Z-matrix of Geometric Parametersa (Bond in Å, Angle and Torsion in degree) for the HF/6-31G* Fully-Optimized Minimum-Energy Structures of DP, MDP, and TP.
Diphosphate (stg I)
P2-O1 1.5326 O3-P2-O1 107.0 P4-O3-P2-O1 90.1
O3-P2 1.6668 P4-O3-P2 159.3 O5-P4-O3-P2 90.1
P4-O3 1.6668 O5-P4-O3 107.0 O6-P2-O3-P4 -151.5
O5-P4 1.5326 O6-P2-O3 104.7 O7-P2-O3-P4 -31.6
O6-P2 1.5359 O7-P2-O3 108.9 O8-P4-O3-P2 -31.6
O7-P2 1.5287 O8-P4-O3 108.9 O9-P4-O3-P2 -151.5
O8-P4 1.5287 O9-P4-O3 104.7 O9-P4 1.5359
Methyl diphosphate gg I
P2-O1 1.5058 O3-P2-O1 104.4 P4-O3-P2-O1 83.2
O3-P2 1.7608 P4-O3-P2 149.3 O5-P4-O3-P2 84.0
P4-O3 1.5635 O5-P4-O3 102.4 C6-O5-P4-O3 -67.6
O5-P4 1.6799 C6-O5-P4 116.8 H7-C6-O5-P4 43.1
C6-O5 1.3906 H7-C6-O5 112.1 H8-C6-O5-P4 -79.1
H7-C6 1.0844 H8-C6-O5 111.2 H9-C6-O5-P4 163.4
H8-C6 1.0943 H9-C6-O5 108.0 O10-P2-O3-P4 -157.1
H9-C6 1.0992 O10-P2-O3 101.3 O11-P4-O3-P2 -29.8
O10-P2 1.5094 O11-P4-O3 114.8 O12-P2-O3-P4 -38.0 O11-P4 1.4923 O12-P2-O3 104.3 O11-P4-O3-P2 -166.7 O12-P2 1.5139 O13-P4-O3 110.7
O13-P4 1.4941 cg
P2-O1 1.5159 O3-P2-O1 102.9 P4-O3-P2-O1 78.3
O3-P2 1.7600 P4-O3-P2 150.6 O5-P4-O3-P2 -27.0
P4-O3 1.5634 O5-P4-O3 104.2 C6-O5-P4-O3 -57.4
O5-P4 1.6693 C6-O5-P4 117.5 H7-C6-O5-P4 55.9
C6-O5 1.3905 H7-C6-O5 111.6 H8-C6-O5-P4 -66.8
H7-C6 1.0836 H8-C6-O5 111.1 H9-C6-O5-P4 175.6
H8-C6 1.0932 H9-C6-O5 107.9 O10-P2-O3-P4 -163.0
H9-C6 1.0980 O10-P2-O3 101.6 O11-P4-O3-P2 -139.7 O10-P2 1.5082 O11-P4-O3 109.8 O12-P2-O3-P4 -42.2
O11-P4 1.5003 O12-P2-O3 105.4 O13-P4-O3-P2 86.0
gg II
P2-O1 1.5076 O3-P2-O1 101.7 P4-O3-P2-O1 156.6
O3-P2 1.7625 P4-O3-P2 149.4 O5-P4-O3-P2 50.1
P4-O3 1.5649 O5-P4-O3 103.8 C6-O5-P4-O3 -54.4
O5-P4 1.6722 C6-O5-P4 118.0 H7-C6-O5-P4 77.2
C6-O5 1.3930 H7-C6-O5 111.2 H8-C6-O5-P4 -43.9
H7-C6 1.0816 H8-C6-O5 111.6 H9-C6-O5-P4 -162.7
H8-C6 1.0850 H9-C6-O5 107.8 O10-P2-O3-P4 -84.5
H9-C6 1.1012 O10-P2-O3 102.9 O11-P4-O3-P2 -64.7
O10-P2 1.5134 O11-P4-O3 113.7 O12-P2-O3-P4 35.7
O11-P4 1.4947 O12-P2-O3 105.1 O13-P4-O3-P2 160.5 O12-P2 1.5068 O13-P4-O3 110.8
O13-P4 1.4950 gt
P2-O1 1.5068 O3-P2-O1 103.0 P4-O3-P2-O1 86.6
O3-P2 1.7894 P4-O3-P2 148.5 O5-P4-O3-P2 86.6
P4-O3 1.5438 O5-P4-O3 100.3 C6-O5-P4-O3 -168.3
O5-P4 1.7023 C6-O5-P4 115.9 H7-C6-O5-P4 64.5
C6-O5 1.3688 H7-C6-O5 112.4 H8-C6-O5-P4 -57.0
H7-C6 1.0888 H8-C6-O5 112.5 H9-C6-O5-P4 -175.7
H8-C6 1.0943 H9-C6-O5 109.0 O10-P2-O3-P4 -154.7
H9-C6 1.0988 O10-P2-O3 100.6 O11-P4-O3-P2 -25.8
O10-P2 1.5090 O11-P4-O3 117.5 O12-P2-O3-P4 -34.9 O11-P4 1.4924 O12-P2-O3 104.8 O13-P4-O3-P2 -163.5 O12-P2 1.5040 O13-P4-O3 111.3
O13-P4 1.4998
Triphosphate
P2-O1 1.5219 O3-P2-O1 108.7 P4-O3-P2-O1 0.0 O3-P2 1.7076 P4-O3-P2 146.0 O5-P4-O3-P2 180.0 P4-O3 1.6309 O5-P4-O3 100.3 O6-P2-O3-P4 120.9 O5-P4 1.6309 O6-P2-O3 105.0 O7-P2-O3-P4 -120.9 O6-P2 1.5280 O7-P2-O3 105.0 O8-P4-O3-P2 64.6 O7-P2 1.5280 O8-P4-O3 109.7 O9-P4-O3-P2 -64.6 O8-P4 1.5017 O9-P4-O3 109.7 P10-O5-P4-O3 180.0 O9-P4 1.5017 P10-O5-P4 146.0 O11-P10-O5-P4 0.0 P10-O5 1.7076 O11-P10-O5 108.7 O12-P10-O5-P4 -120.9 O11-P10 1.5219 O12-P10-O5 105.0 O13-P10-O5-P4 120.9 O12-P10 1.5280 O13-P10-O5 105.0
O13-P10 1.5280
[image:3.612.101.508.83.588.2]a see Figure S7 for atom labeling.
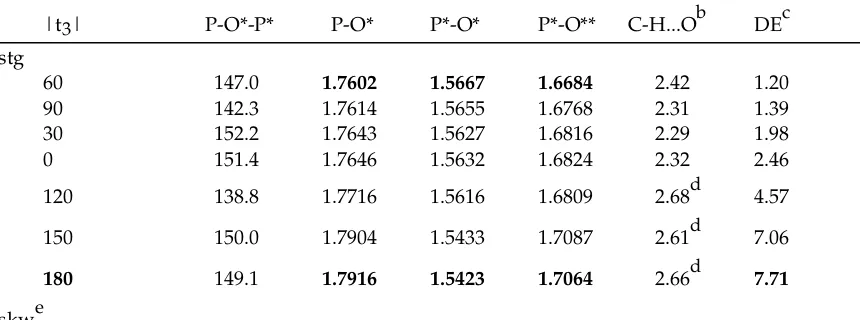
TABLE S2: Conformational Energy Differences (DE, in kcal/mol) and Selected Geometric Parametersa (Bond in Å, Angle and Torsion in degree) for the HF/6-31G* Optimized Phosphate Rotational Isomers of Methyl Diphosphate (MDP).
stg
60 150.7 1.7591 1.5623 1.6755 2.16 0.18
90 148.3 1.7611 1.5633 1.6825 2.24 0.27
30 152.1 1.7581 1.5618 1.6707 2.15 0.28
0 151.7 1.7587 1.5624 1.6683 2.18 0.32
120 148.0 1.7632 1.5607 1.6928 2.54 1.24
150,180 180.0 1.7604 1.5432 1.6897 2.64 2.50 skwd
90 I 150.1 1.7623 1.5634 1.6814 2.16 0.28
60 I 153.2 1.7603 1.5614 1.6747 2.10 0.46
30 I 152.7 1.7609 1.5625 1.6700 2.13 0.60
60 II 150.7 1.7623 1.5628 1.6767 2.24 0.81
30 II 158.1 1.7593 1.5577 1.6724 2.11 0.82
0 157.8 1.7597 1.5582 1.6710 2.12 1.30
120 I 152.9 1.7627 1.5579 1.6916 2.43 1.32
90 II 151.4 1.7669 1.5604 1.6856 2.55 1.74
120 II,150,180 180.0 1.7581 1.5459 1.6866 2.41 1.96 eclp
60 153.1 1.7623 1.5621 1.6749 2.16 0.97
90 153.1 1.7647 1.5606 1.6834 2.26 1.21
30 157.8 1.7611 1.5585 1.6717 2.15 1.29
0 159.8 1.7613 1.5570 1.6714 2.16 1.44
120 160.1 1.7642 1.5525 1.6916 2.50 2.26
150,180 180.0 1.7602 1.5466 1.6866 2.37 2.37
a see Figure 1 for the labeling of atoms and torsions.
b the shortest C-H...O distance.
c
with respect to the fully-optimized minimum of lowest energy (gg I, Table II). d I and II represent two different structures with the same absolute value of t
[image:4.612.91.521.541.701.2]2.
TABLE S3: Conformational Energy Differences (DE, in kcal/mol) and Selected Geometric Parametersa (Bond in Å and Angle and Torsion in degree) for the HF/6-31G* Optimized Methyl Rotational Isomers of Methyl Diphosphate (MDP)
|t3| P-O*-P* P-O* P*-O* P*-O** C-H...Ob DEc
stg
60 147.0 1.7602 1.5667 1.6684 2.42 1.20
90 142.3 1.7614 1.5655 1.6768 2.31 1.39
30 152.2 1.7643 1.5627 1.6816 2.29 1.98
0 151.4 1.7646 1.5632 1.6824 2.32 2.46
120 138.8 1.7716 1.5616 1.6809 2.68d 4.57
150 150.0 1.7904 1.5433 1.7087 2.61d 7.06
180 149.1 1.7916 1.5423 1.7064 2.66d 7.71
60 I 149.2 1.7618 1.5637 1.6820 2.15 0.20
90 I 143.9 1.7598 1.5655 1.6747 2.22 0.96
60 II 147.0 1.7620 1.5666 1.6690 2.50 1.13
30 I 150.1 1.7649 1.5642 1.6782 2.22 1.63
30 II 150.9 1.7649 1.5627 1.6879 2.16 2.03
90 II 141.8 1.7626 1.5650 1.6741 2.39 2.42
0 150.6 1.7636 1.5635 1.6802 2.44 2.79
120 I 139.2 1.7728 1.5606 1.6818 2.38d 5.13
120 II 139.3 1.7715 1.5614 1.6783 2.38d 5.23
150 I 146.5 1.7853 1.5477 1.6973 2.52d 6.54
180 149.1 1.7913 1.5424 1.7055 2.52d 7.07
150 II 146.5 1.7858 1.5472 1.6978 2.33d 7.65 eclp
60 150.1 1.7602 1.5638 1.6686 2.21 0.32
30 150.2 1.7638 1.5634 1.6784 2.23 1.69
90 143.6 1.7609 1.5650 1.6723 2.35 1.97
0 149.6 1.7672 1.5627 1.6851 2.31 3.46
120 139.5 1.7727 1.5605 1.6788 2.25d 5.67
180 149.2 1.7907 1.5427 1.7045 2.65d 6.62
150 146.6 1.7856 1.5474 1.6976 2.31d 7.19
a see Figure 1 for the labeling of atoms and torsions.
b the shortest C-H...O distance.
c with respect to the fully-optimized minimum of lowest energy (gg I, Table II).
d where the oxygen belongs to the phosphate group adjacent to the methyl.
[image:5.612.92.522.565.676.2]e I and II represent two different structures with the same absolute value of t 3.
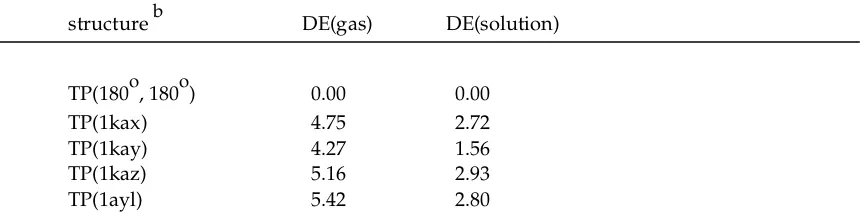
Table S4. Conformational Energy Differences (DE, in kcal/mol) of TP Structures Optimized at Protein-Bound ATP Torsional Angles of Its Pyrophosphate Bonds, in Gas Phase and in Solution Phasea
structure b DE(gas) DE(solution)
TP(180o, 180o) 0.00 0.00
TP(1kax) 4.75 2.72
TP(1kay) 4.27 1.56
TP(1kaz) 5.16 2.93
a The gas phase structures were torsion-constrained-optimized with HF 6-31G* using
Gaussian94,17 while the solution phase results were obtained by including the solvation effect according to Jaguar18,19 on the gas-phase structures. The Jaguar parameters used are molchg = -5, multip = 1, isolv = 2, igeopt = 0, and basis = 6-31G*.
b The reference is a gas-phase global minimum whose structure is depicted in Figure S5. During