David Publishing Company www.davidpublishing.com P u b l i s h i n g Dav i d
Journal of Life Sciences
Publication Information
Journal of Life Sciences is published monthly in hard copy (ISSN 1934-7391) and online (ISSN 1934-7405) by David Publishing Company located at 1840 Industrial Drive, Suite 160, Libertyville, Illinois 60048, USA.
Aims and Scope
Journal of Life Sciences, a monthly professional academic journal, covers all sorts of researches on molecular biology, microbiology, botany, zoology, genetics, bioengineering, ecology, cytology, biochemistry, and biophysics, as well as other issues related to life sciences.
Editorial Board Members
Dr. Stefan Hershberger (USA), Dr. Suiyun Chen (China), Dr. Farzana Perveen (Pakistan), Dr. Francisco Torrens (Spain), Dr. Filipa João (Portugal), Dr. Masahiro Yoshida (Japan), Dr. Reyhan Erdogan (Turkey), Dr. Grzegorz Żurek (Poland), Dr. Ali Izadpanah (Canada), Dr. Barbara Wiewióra (Poland), Dr. Valery Lyubimov (Russia), Dr. Amanda de Moraes Narcizo (Brasil), Dr. Marinus Frederik Willem te Pas (The Netherlands), Dr. Anthony Luke Byrne (Australia), Dr. Xingjun Li (China), Dr. Stefania Staibano (Italy), Dr. Wenle Xia (USA), Hamed Khalilvandi-Behroozyar (Iran).
Manuscripts and correspondence are invited for publication. You can submit your papers via Web Submission, or E-mail to [email protected] or [email protected]. Submission guidelines and Web Submission system are available at http://www.davidpublishing.com.
Editorial Office
1840 Industrial Drive, Suite 160, Libertyville, Illinois 60048 Tel: 1-847-281-9862, Fax: 1-847-281-9855
E-mail:[email protected], [email protected]
Copyright©2011 by David Publishing Company and individual contributors. All rights reserved. David Publishing Company holds the exclusive copyright of all the contents of this journal. In accordance with the international convention, no part of this journal may be reproduced or transmitted by any media or publishing organs (including various websites) without the written permission of the copyright holder. Otherwise, any conduct would be considered as the violation of the copyright. The contents of this journal are available for any citation. However, all the citations should be clearly indicated with the title of this journal, serial number and the name of the author.
Abstracted / Indexed in
Database of EBSCO, Massachusetts, USA Chemical Abstracts Service (CAS), USA Cambridge Scientific Abstracts (CSA), USA
Chinese Database of CEPS, American Federal Computer Library center (OCLC), USA Ulrich’s Periodicals Directory, USA
Chinese Scientific Journals Database, VIP Corporation, Chongqing, China
Subscription Information
Price (per year): Print $420, Online $300, Print and Online $560
David Publishing Company
1840 Industrial Drive, Suite 160, Libertyville, Illinois 60048 Tel: 1-847-281-9862, Fax: 1-847-281-9855
E-mail: [email protected]
Dav id Publishing Company w ww.davidpublis hing.com
Pu b li sh i ng
Dav i d
J LS
Journal of Life Sciences
Volume 5, Number 9, September 2011 (Serial Number 41)
Contents
Research Papers
677 Statistical Analysis of Genetic Diversity in 15 STR Loci from Han, Miao and Yao Tribes in South China
Kuanheng Wu, Miao He, Yijing Wang, Jian Song, Liping Ling and Daniel Wai Tin Chan
682 CCR5-CCR2 Gene Polymorphisms in Ethiopian Jews: Population Divergence and Its Relevance
to HIV-1 Infection Resistance
Michael Korostishevsky, Batsheva Bonne-Tamir, Zvi Bentwich, Alexander Kalinkovich and Alexander
Tsimanis
690 First Record of Frankliniella Occidentalis and Impatiens Necrotic Spot Virus in Egypt
Abeer Salah El-Deen Abd El-Wahab, Mohamed Abdel-Kader El-Sheikh and Salah Elnagar
697 Investigation of the Optimum Condition and Antimicrobial Activities of Pigments from Four
Potent Pigment-Producing Fungal Species
Neveen S. Geweely
712 Effect of Ginger Powder (Zingiber Officinale) on Plasma Lipid Profile and Liver Enzyme
Activities of Hypercholesterolemic Rats
Ajayi Olubunmi Bolanle
717 Parasitic Plants as a New Target Plant for Screening Rice Allelopathic Potential
Yiqing Guo, Kil-Ung Kim, John I. Yoder and Donghyun Shin
725 Effect of Graded Levels of Nitrogen on Growth and Yield of Eggplant (Solanum Melongena) in Kabba, Southern Guinea Savanna Ecological Zone of Nigeria
John Akintola Oloniruha
728 Alternative Technologies Used in Laying Hens Husbandry
18S rRNA Sequences
Madhav V. Upadhye, Rajesh C. Patil, Sonal M. Manohar and Ujwala Jadhav
739 Effect of Thermal Stress, Cistern Size and Milking Frequency on Plasma Mineral Concentrations in Holstein Dairy Cows
Rim Ben Younes, Moez Ayadi, Taha Najar, Margherita Caccamo, Iris Schadt and Moncef Ben M’Rad
747 Effect of Ageing Time on Meat Characteristics of Castrated and Uncastrated Brahman Cattle
José A. Miguel, Jesús Ciria, Begoña Asenjo, David Colmenarez and Hector Pargas
754 Effect of Climate Change on Phenology of Forage Grass Species
Żurek Grzegorz
759 Developing Adaptation Strategies Due to Climate Change: With Special Reference to the
Vulnerable Java Fisheries, Indonesia
Indah Susilowati and Agus Hartoko
768 Fast Determination of Cd, Pb, and Cu in Grape Must and Wine
Jarmila Lastincova, Ernest Beinrohr and Lubica Pospísilova
772 Effect of the Chromatic Assimilation (Bezold Effect) in the Vision of the Content on a Dinner Plate
Statistical Analysis of Genetic Diversity in 15 STR Loci
from Han, Miao and Yao Tribes in South China
Kuanheng Wu1, Miao He 1, Yijing Wang1, Jian Song1, Liping Ling2 and Daniel Wai Tin Chan3
1. Life Sciences School, Sun Yat-sen University, Guangzhou, Guangdong 510275, China
2. Education School, Sun Yat-sen University, Guangzhou, Guangdong 510275, China
3. Department of Building Services Engineering, The Hong Kong Polytechnic University, Hunghom, Hong Kong, China
Received: August 18, 2010 / Accepted: November 03, 2010 / Published: September 30, 2011.
Abstract: It is interesting to find the possible statistical characters of 15 STRs in Han, Miao and Yao, the three main tribal populations
in South China, and the significant differences of allele frequencies by comparing STR loci in each cluster for applications of forensic science. Genetic diversity in 15 STR loci [D8S1179, D21S11, D7S820, CSF1PO, D3S1358, TH01, D13S317, D16S539, D2S1338, D19S433, vWA, TPOX, D18S51, D5S818, and FGA] from Han, Miao and Yao tribes in South China had been analyzed. The allele frequencies of 15 tetrameric STR loci were obtained from 1,530 unrelated individuals of three main tribal populations [Han, Yao and Miao] inhibiting in South China. Cluster analysis and LSD test had been used for data analysis. The high degree statistical differentia of genetic polymorphism has been found among three tribal populations.
Key words: STR, genetic diversity, statistics, Han, Miao, Yao.
Abbreviations: STR: short tandem repeats; The traditional STR loci applied in forensic science includes 15 STR loci [D8S1179, D21S11, D7S820, CSF1PO, D3S1358, TH01, D13S317, D16S539, D2S1338, D19S433, vWA, TPOX, D18S51, D5S818, and FGA]; LSD: a least significant difference test of one-way analysis of variance.
1. Introduction
15 STR loci from three tribal populations Han, Miao and Yao in South China were obtained in present study. Whole DNA extracted from venous blood was obtained from randomly unrelated 1,000 Han, 248 Yao and 282 Miao individuals, Guangdong Province. Tribe Miao and Yao have over nine million people inhabiting mainly in mountainous areas of Guizhou, Guangxi and Hainan Province.
The traditional methods of Hardy-Weinberg equilibrium, the power of discrimination, the probability of paternity exclusion, and the polymorphic information content have already presented profound conclusions about the significant
Corresponding author: Miao He, Ph.D., associate professor,
research fields: bioinformatics, biostatistics. E-mail: [email protected].
the genetic polymorphism presented by cluster analysis especially the genetic diversity between Han and two other tribes; (2) a region of allele frequencies selected by LSD test revealing significant differences among STR loci in the same cluster could serve as a genetic marker for individual identification among three tribal populations; (3) collecting the genetic polymorphic information from the three tribal populations and retrospectively interpreting the demographic tribal migration and amalgamation in South China.
2. Materials and Methods
2.1 DNA Analyzed
The allele distributions of 15 STR loci were obtained from 1,530 unrelated individuals [2-6]. Genome DNA was extracted from whole blood using Chelex-100 extraction method (Bio-lad Company). Those samples failed in former method using standard phenol chloroform methods or DNA IQTM commercial kits (Promega Company) extract DNA again [7].
2.2 STR Typing
All samples were PCR amplified using AmpFlSTR® Identifiler™ commercial kits (Applied Biosystems Company) [8]. PCR amplifications were carried out according to manufacturers (AmpF/STR Indetifi-ler PCR amplification kit, AB Applied Biosystems). Amplification products were run in an ABIPRISM 3100 genetic analyzer (ABI Company) using Liz500 as internal standard label. Genescan 3.7 (ABI Company) software was used to collect the data, analyze fragment sizes and Genepop v3.4 to calculate the allele frequencies of each STR locus in 3 tribes [9].
2.3 Statistical Analysis
Cluster analysis was applied on 15 STR loci from each tribal population using the correlation matrix calculated from 15 sets of STR loci allele frequencies [10, 11]. By comparing each pair of three cluster results, three STR loci [D8S1179, D7S820, and D13S317] probably indicated the genetic diversity among three
tribes. Further variance analysis was applied on these STR loci using LSD value, calculated from two sets of STR locus allele frequencies array or mean allele frequencies array of each allele from several STR loci in the same cluster, to discriminate the significant differences of allele frequencies between two STR loci [12, 13]. To be specific, we calculated LSD (= 0.05) value from two arrays {X1, X2, X3 …} and {Y1, Y2,
Y3 …} which represented allele frequencies of STR
locus X and Y separately. If |Xi- Yi | > LSD, the allele
frequencies Yi is significantly different from Xi.
Microsoft Office Excel 2007 and SPSS 15 were used to analyze these data.
3. Result
Cluster analysis preformed on correlation matrix
indicated several profound conclusions which were then further studied by using LSD test. The results of hierarchical cluster analysis revealed highly
consistent clusters iv among three tribal populations and clusters [i, ii, iii] shared genetic diversity to variant extend. LSD test applied on three STR loci
[D8S1179, D7S820, D13S317] of Han further corroborated the significant diversity of clusters [i,
iii] among Han, Miao and Yao.
3.1 Cluster Analysis
Fig. 1 The cluster analysis of Han (a), Miao (b) and Yao (c). The dissimilarities focused mainly on clusters [i, ii, iii], rather than iv were exactly the same among three tribes. The topologic structures in cluster iii varied between Han and Miao, Han and Yao.
3.2 Variance Analysis
LSD values were used to discriminate the significant differences of allele frequencies from specific STR loci [D8S1179, D7S820, and D13S317] of Han selected by cluster analysis above between each pair of tribes.
3.2.1 LSD Test between Han D8S1179 and STR Loci [D3S1358, vWA, D18S51, D19S433] from both Miao and Yao
Since Han’s D8S1179 had the similar hierarchy of cluster analysis to Miao and Yao’s STR loci [D3S1358, vWA, D18S51, D19S433], the result in Table 1 revealed the significant differences allele frequencies ranging from allele 10 to 18 in four STR loci of Miao
and Yao. A set of symbols “-”, “+--”, “+-”, “+”, “++”, “+++”, “++++” were used to represent the levels of significant difference (< 0], (0, 0.04], (0.04, 0.08], (0.08, 0.12], (0.12, 0.16], (0.16, 0.20], (> 0.20). The “++++” and “+++” symbols focused on allele 13, 16, 17, 18 of STR loci D3S1358 and vWA interpreted a genetic diversity in Han’s D8S1179 and STR loci [D3S1358, vWA, D18S51, D19S433] from both Miao and Yao. By comparison, symbols in two columns of each STR locus indicated the genetic diversity between Miao and Yao. The distinctness located on 5 allele of D19S433 and allele 13 of D3S1358 and vWA.
3.2.2 LSD Analysis between Han’s D7S820 and STR Loci [CSF1PO, D5S818, D16S539, D13S317] from both Miao and Yao
The levels of significant differences about allelic with Han, focused on allele 8. CSF1PO column contained different symbols in two allelic positions of Miao and Yao probably interpreted STR diversity though not significant. Frequencies analyzed in Table 2 were not as distinct as in Table 1.
Table 1 shows that a slot located on allele 8 and several “+” separated in allele 9, 11 and 12 of four STR loci represented the genetic diversity in Han’s D7S820 and four STR loci of Miao and Yao, which had similar hierarchy of cluster analysis with Han, focused on allele 8. CSF1PO column contained different symbols in two allelic positions of Miao and Yao probably interpreted STR diversity though not significant.
3.2.3 LSD Test between Han’s D13S317 and [CSF1PO, D5S818, D16S539, D7S820] from both Miao and Yao
The LSD test results illustrated in Table 3 represented more significant differences compared with Table 2. “++++” symbols mainly located on allele 8 and 12 of four STR loci consolidated the similar results found in Table 2. The deviation information combining Table 2 and Table 3 suggested the most distinctive genetic character between Han [D13S317, D7S820] and [CSF1PO, D5S818, D16S539, D7S820] from Miao and Yao, focused on allele [8, 11, 12]. (b)
Table 1 Han’s D8S1179 with Miao and Yao [D3S1358, vWA, D18S51, D19S433] LSD test.
Allele D3S1358 vWA D18S51 D19S433 Miao Yao Miao Yao Miao Yao Miao Yao Miao) = 0.032820, LSD (Han and Yao) = 0.032858.
Table 2 Han’s D7S820 with Miao and Yao [CSF1PO, D5S818, D16S539 and D7S820].
Allele
Table 3 Han D13S317 with Miao and Yao [CSF1PO, D5S818, D16S539 and D7S820].
Allele CSF1PO D5S818 D16S539 D7S820 Miao Yao Miao Yao Miao Yao Miao Yao
Huge number of STR loci in unrelated three tribal populations of South China has been typed in order to obtain genetic characteristics of these three tribes. Cluster analysis and LSD test have been applied to discover several genetic hypotheses about the tribal differentiation. The most distinctive feature of present study is the genetic diversity among three tribes and several regions of allelic significant [17-20].
The profound conclusions obtained from present study include several points below.
The cluster analysis indicates an exactly identical cluster ⅳ of each tribal population in both members and topological structure. Four STR loci [D2S1138, FGA, D21S11, TH01] included in cluster ⅳ indicate high consistent genetic characters.
Members in clusters [iii, i] vary in three tribal populations, also have the same of topological structure.
The hierarchical positions of the three STR loci [D8S1179, D7S820, and D13S317] vary among three tribes according to cluster analysis.
The statistical analysis reveals the variance between Han’s D8S1179 and clusters ⅲ [D3S1358, vWA, D18S51, D19S433] in Miao and Yao focuses on allele [13, 16, 17, 18] of D3S1358 and vWA.
The variance between Han’s [D13S317, D7S820] and cluster i [CSF1PO, D5S818, D16S539, D7S820] in Miao and Yao focuses on allele [8, 11, 12].
The weightiness of STR loci in cluster ⅳ could be omitted or reduced since the indistinguishable genetic diversity was in paternity testing or personal identify among these three tribes.
The weightiness of cluster [iii, i] supposes to be aggravated because of the high degree of genetic diversity.
Acknowledgment
and Science and Technology Planning Project of Guangdong Province of China (No. 2003A3080503).
References
[1] H.J. Zhuang, Y.B. Li, J.P. Jiang, J. Zhang, J. Wu, H. Du, et al., Analysis of 15 STR loci in Chinese population from Sichuan in West China, Forensic Science International 171 (2007) 222-225.
[2] C. Liu, C.H. Liu, H.J. Wang, Genetic diversity at 15 STR loci in two tribal populations in Southern China, Forensic Science international 162 (2006) 28-32.
[3] J.G. Hirschfeld, M.J. Farfan, V. Prieto, M.L. Soto, Y. Torres, P. Sanz, Allele distribution of 15 STRs in a population from Extremadura (Central-Western Spain), International Congress Series 1239 (2003) 165-169. [4] L.A. Zhivotovsky, S. Ahmed, W. Wang, A.H. Bittles,
The forensic DNA implications of genetic differentiation between endogamous communities, Forensic Science International 119 (2001) 269-272.
[5] M.S. Shi, J.P. Tang, R.F. Bai, X.J. Yu, J.Y. Lv, B. Hu, Haplotypes of 20 Y-chromosomal STRs in a population sample from southeast China (Chaoshan area), Int. J. Legal. Med. 121 (2007) 455-462.
[6] L.A. Zhivotovsky, V.M. Veremeichyk, A.I. Mikulich, I.G. Udina, L.A. Atramentova, S.A. Kotova, et al., A comprehensive population survey on the distribution of STR frequencies in Belarus, Forensic Science Interational 172 (2007) 156-160.
[7] U.D. Immel, M. Krawczak, J. Udolph, A. Richter, H. Rodig, M. Kleiber, et al., Y-chromosomal STR haplotype analysis reveals surname-associated strata in the East-German population, European Journal of Human Genetics 14 (2006) 577-582.
[8] P.S. Walsh, D.A. Metzger, R. Higuchi, Chelex 100 as a medium for simple extration of DNA for PCR-based typing from forensic material, Biotechniques 10 (1991) 506-513.
[9] P. Wiegand, T. bajanowski, B. Brinkmann, PCR typing
of debris from fingemails, Int. J. Legal. Med. 106 (1993) 81-84.
[10] M. Raymond, F. Rousset, GENEPOP: Populations genetics software for exact tests and ecumenicism, Journal of Heredity 86 (1995) 248-249.
[11] C.C. Cockerham, Analyses of gene frequencies, Genetics 74 (1973) 679-700.
[12] D.J. Balding, R.A. Nichols, DNA profile match probability calculation: How to allow for population stratification relatedness, database selection and single bands, Forensic Sci. International 64 (1994) 125-140. [13] S. Schneider, J.M. Kueffer, D. Roessli, L. Excoffier, A
Softerware for Population Genetic Data Analysis, Genetics and Biometry Laboratory, University of Geneva, Switzerland, 1997.
[14] J. Goudet, M. Raymond, T.D. Meeus, F. Rousset, Test differentiation in diploid populations, Genetics 144 (1996) 1993-1940.
[15] S.W. Guo, E.A. Thompson, Performing the exact test of Hardy-Weinberg proportions for multiple alleles, Biometrics 48 (1992) 361-372.
[16] J.B.S. Haldane, An exact test for randomness of mating, Journal of Genetics 52 (1954) 631-635.
[17] E.J. Louis, E.R. Dempster, An exact test for Hardy-Weinberg and multiple alleles, Biometrics 43 (1987) 805-811. [18] E. Bosch, F. Calafell, A.P. Lezaun, J. Clarimon, D.
Comas, E. Mateu, et al., Genetic structure of north-west Africa revealed by STR analysis, European Jouranl of Human Genetics 8 (2000) 360-366.
[19] L. Garofano, M. Pizzamiglio, C. Vecchio, Italian population data in thirteen short tandem repeat loci: HUMTHO1, D21S11, D18S51, HUMVWFA31, HUMFIBRA, D8S1179, HUMTPOX, HUMCSF1PO, D16S539, D7S820, D13S317, D5S818, D3S1358, Forensic Science International 97 (1998) 53-60.
CCR5-CCR2 Gene Polymorphisms in Ethiopian Jews:
Population Divergence and Its Relevance to HIV-1
Infection Resistance
Michael Korostishevsky1, Batsheva Bonne-Tamir2, Zvi Bentwich3, Alexander Kalinkovich4 and Alexander Tsimanis5
1. Department of Anatomy and Anthropology, Tel Aviv University, Tel Aviv 69978, Israel
2. Department of Human Molecular Genetics & Biochemistry, Tel Aviv University, Tel Aviv 69978, Israel 3. Rosetta Genomics, Rehovot 76706, Israel
4. Department of Immunology, Weizmann Institute of Science, Rehovot 76100, Israel 5. Biona Ltd, Rehovot 76120, Israel
Received: January 25, 2011 / Accepted: May 18, 2011 / Published: September 30, 2011.
Abstract: CCR5 and CCR2 genes have been implicated in HIV disease progression and HIV resistance in various human populations
but not in the Ethiopian Jews. The authors examined polymorphisms in the CCR5-CCR2 gene region in two groups of Ethiopian Jews, 29 non-exposed and 13 exposed but uninfected individuals. Prevalence of the CCR2-V64I and CCR5-32 mutations as well as genetic variations in the CCR5 promoter region at positions 208, 627, 676 and 927 has been studied. The authors confirmed the absence of CCR5-32 mutation in all individuals studied. Three CCR5 single-nucleotide polymorphisms (SNPs) G208T, T627C and A676G were in tight linkage disequilibrium (LD) with each other. In contrast, a lack of LD was observed across the above-mentioned SNPs and proximal SNPs C927T and distal CCR2-G190A. Only four CCR5 haplotypes - HHA, HHC, HHE and HHF*2 were identified in both groups. Using multi-SNP analysis, no significant differences in the genotype frequencies between the groups were found (χ23df = 4.66,
P = 0.198). Observed deviation in a single SNP allele frequency (T627C SNP: χ21df = 4.14, P = 0.042) was not preserved after the Bonferroni correction. Allelic frequencies were compared to other geographically targeted worldwide populations, where clear distinction between Ethiopian Jews and Africans has been found. These data were reflected in the phylogenetic tree, in which Ethiopian Jews branch with Asians.
Key words: CCR5 and CCR2 polymorphism, Ethiopian Jews, HIV-1 infection resistance.
1. Introduction
The observation that chemokine receptors are used by HIV as coreceptor for the cellular entry led to the discovery of genetic factors that can affect susceptibility to infection with HIV or the rate of progression to disease once infection is established [1, 2]. Functionally important polymorphisms in the regulatory region of CCR5 gene, a 32-base pair deletion in the coding part
Corresponding author: Michael Korostishevsky, Ph.D.,
senior scientist, research field: genetic epidemiology. E-mail: [email protected].
CCR5-32 deletion is the best characterized genetic trait. In the epidemiological studies, the allelic frequency of the deletion was 10%-20% among Caucasians, particularly amongst those of Northen European descent with 1% homozygosity. This mutation is extremely rare in African and Asian population [8, 9]. Individuals homozygous for CCR5-32 mutation are almost completely resistant to HIV infection most probably due to lack of CCR5 receptor on their cell surfaces [10]. Studies of CCR5-32 mutation in exposed but uninfected individuals have revealed that only a small proportion of them were homozygous for this mutation [11]. Heterozygosity for CCR5-32 mutation is associated with delayed progression to AIDS in infected individuals. Moreover, frequency of heterozygosity is significantly higher in long term non-progressors than in progressors and rapid progressors [8-10, 12, 13]. The mechanism of protection is not clear and it is believed that CCR5 expression may be altered in these individuals. CCR2-V64I mutation is associated with delay in progression to AIDS, probably due to the heterodimerization and sequestration of the CCR5 receptor [14, 15].
Our research focused on CCR5 genetics of Ethiopian Jews currently living in Israel. This population originated in the north of Lake Tana in Gondar, Ethiopia, and several thousands of them were airlifted to Israel first during the Ethiopian civil war (1984-1985) and then in 1989.
The purpose of this study was to examine whether the DNA polymorphisms at the loci that encode CCR5 and CCR2 receptors, can potentially explain the persistent seronegativity in a group of exposed seronegative (ESN) individuals. Using evolutionary-based CCR5 haplotype classification [3, 16], the authors have characterized this polymorphism in two groups of Ethiopian Jews: Healthy individuals without any history of HIV infection or ESN individuals. The authors assessed of CCR5-32 deletion as well as several single nucleotide polymorphisms (SNPs) in the CCR5 promoter region and coding part
of CCR2 gene. The authors estimated the magnitude of linkage disequilibrium between the SNPs and performed multi-SNP analysis between the samples. Using the CCR5 haplotype distribution data for different ethnic groups, the authors investigated the phylogenetic relationships for Ethiopian Jews.
2. Materials and Methods
2.1 Subjects
A total of 29 HIV-1-negative (control) and 13 exposed but uninfected seronegative (ESN) Ethiopian Jews were sampled for this study. The evaluation of clinical status of individuals including the presence of antibodies to HIV-1 and HIV-1 viral particles has been performed in the Kaplan Medical Center, Rehovot, Israel, as described [17]. Samples were coded and tested blind. Informed consent was obtained for the collected samples.
2.2 Genotyping
The authors verified the presence of CCR5-Δ32 deletion and genotyped sequence variations for the single-nucleotide polymorphism (SNP) G208T, T627C, A676G and C927T in the CCR5 promoter region as well as G190A mutation in the coding part of CCR2 gene. The authors used genomic DNA obtained from peripheral blood lymphocytes. The DNA samples were subjected to a polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) assay as previously described [12, 18, 19]. PCR amplification was performed to amplify CCR5 promoter and CCR5 and CCR2 genes fragments covering the polymorphic sites (Table 1).
For CCR2-V64I mutation, the PCR products were digested with FokI. After digestion, the products of digestion for CCR2 and the PCR products for delta-32 CCR5 were loaded on a 4% agarose gel in 0.5 × TBE and electrophoresed at 85 V for 2 hours.
Table 1 SNPs and primer to PCR-amplify the CCR5 and CCR2 genes.
SNP Position* Primer Sequence *The numbering of nucleotide positions is based on GenBank sequence U95626.
were resolved by electrophoresis in 6.5% polyacrylamide gel.
2.3 Statistical Analysis
Genotype and allele frequencies of the SNPs were calculated by direct counting. Possible differences in the frequencies of each of the SNP genotypes and alleles between the samples were estimated using the χ2 test, as described elsewhere [20, 21]. The Arlequin software package [22] was used for: 1) Evaluation of genetic distances between different populations [23]; 2) Estimation of pairwise LD between the SNP markers [24, 25]; 3) Detection of departure from Hardy-Weinberg equilibrium (HWE) [26]; and 4) Calculation of the maximum likelihood (ML) of haplotype frequencies [27].
Based on the ML haplotype frequency estimates, the likelihood ratio test (LRT) for sample differentiation was evaluated as previously described [28]. PHYLIP software package [29] was used for phylogeny inferences based on the CCR2-CCR5 region genetic distances. The Bonferroni corrections were performed by the SISA online procedure (http://home.clara.net/sisa/bonfer.htm).
3. Results and Discussion
3.1 Genotype and Allele Frequencies
Both the control and ESN groups were genotyped for the CCR5-32, CCR2-G190A and CCR5 promoter alleles constituting the CCR5 human haplotypes.
The authors performed DNA PCR by use of primers
that amplified the region encoding the 32-bp deletion. CCR5-32 deletion was not detected in either the control or ESN groups, indicating that this mutant allele is probably rare or absent in the Ethiopian Jewish population. CCR5-32 allele is very common in white populations, yet this allele is rare in people of African and Asian descent including Ethiopian Jews [30].
The results of LD tests between pairs of SNP markers for ESN and control groups are depicted in Fig. 1. As can be seen, three internal SNPs, CCR5-G208T, CCR5-T627C and CCR5-A676G, demonstrated significant linkage disequilibrium (P < 0.05). Distal SNP CCR2-G190A and proximal CCR5-C927T were found in strong linkage disequilibrium (P < 0.0005), while a lack of linkage disequilibrium between them and the internal SNPs was observed. Our results confirmed the strong linkage disequilibrium between CCR2-64I and CCR5-927T, which had been reported previously [31].
Allelic and genotype frequencies for the CCR2-G190A and four CCR5 promoter polymorphisms among the control and ESN individuals are shown in Table 2.
A discrepancy in allelic frequency between the control and ESN individuals was found for the CCR5-A676G SNP (χ21df = 4.14, P = 0.042). The
C927T
A676G
T627C
G208T
V64I
C927T
A676G
T627C
G208T
V64I
SNP
Fig. 1 Significance level for pairwise LD in the ESN and control samples. LD significance above the diagonal corresponds to
the ESN sample and below the diagonal to the control sample (white: P > 0.05; gray: 0.005 < P < 0.05; dark gray: 0.0005 < P <
0.005; black: P < 0.0005).
Table 2 Genotype and allele frequencies of each SNP1 in ESN and control samples.
SNPs Genotype distribution Allele distribution
ID2 Position3 Genotype ESN Control χ2 test Allele ESN Control χ2 test CCR2-64I
(G190A)
46,295 GG 7 17 2.29
G 19 46 0.40
GA 5 12
AA 1 0
A 7 12
CCR5-G208T 58,934 GG 7 8 3.87
G 20 33 3.09
GT 6 17 TT 0 4
T 6 25
CCR5-T627C 59,353 TT 1 8 4.57
T 9 34 4.14
CT 7 18
CC 5 3
C 17 24
CCR5-A676G 59,402 AA 7 8 3.87
A 6 33 3.09
AG 6 17
GG 0 4
G 20 25
CCR5-C927T 59,653 TT 1 0 2.29
C 19 46 0.40
CT 5 12 CC 7 17
T 7 12
1
SNPs that were non-polymorphic in both samples are not presented in the table: CCR5-A29G (100% A) and CCR5-630 (100% C). 2
SNP designations are according to Ref. [32].
3.2 Haplotype Polymorphism
Only four haplotypes among known CCR5 haplotypes were detected in ESN and control individuals (Table 3).
The haplotypes are notated according to the evolutionary-based classification of the CCR5 [3, 16]. The HHF*2 frequency in the ESN individuals is slightly higher than that in the control group (26.9% vs. 20.7%), although the difference did not attain statistical significance. In the control group, the most common CCR5 haplotype was HHC (43.1%), whereas among ESN individuals, the most common haplotype was HHE (38.6%). Of note, both haplotypes have significantly higher frequencies in Caucasians [3]. The minor haplotype in both studied groups was HHA (15.5% and 11.5%, respectively), which is more frequent in Africans. However, the multi-SNP likelihood ratio test did not find significant differences between the studied groups (χ23df = 4.66, P = 0.198). Interestingly,
genotyping of samples from both studied groups failed to detect presence of HHD and/or HHB haplotypes, which are specific to African population [3, 16].
Eight different genotypes were found in the control group: HHA/HHA, HHA/HHC, HHA/HHF*2, HHA/HHE, HHC/HHC, HHC/HHE, HHC/HHF*2, HHE/HHF*2. In the ESN group seven haplotype pair combinations, namely HHA/HHC, HHA/HHF*2, HHA/HHE, HHC/HHE, HHC/HHF*2, HHE/HHE and HHF*2/HHF*2 were detected. In the control group, the most prevalent haplotype pairs were HHC/HHF2 (27.6%), followed by HHC/HHC and HHC/HHE. On the other hand, in the ESN group the leading haplotype
Table 3 ML estimates of haplotype frequencies in ESN and control samples.
Haplotype HH ESN Control OR LRT*(P-value) G-G-C-A-C HHE 0.385 0.207 1.859
4.66 (0.198) G-T-T-G-C HHC 0.231 0.431 0.535
A-G-C-A-T HHF*2 0.269 0.207 1.301 G-G-T-A-C HHA 0.115 0.155 0.744
* The likelihood ratio test (LRT) for sample differentiation was evaluated as previously described [28].
pairs was HHC/HHE (23.1%), followed by HHC/HHF*2, HHE/HHE and HHE/HHF*2, the haplotypes that were the most common pairs. Such distribution of haplotype pairs, both in the control and ESN groups are typical of Caucasians, whereas two pairs, HHA/HHA and HHA/HHF*2 haplotypes, typical of Africans [32] were rare in Ethiopian Jews.
level of CCR5 was found in HIV-exposed uninfected females and unexposed controls from Kenya and Ethiopia [40, 41]. Taken together, these data suggest that the distribution of polymorphisms in CCR5 promoter region varies significantly across race/ethnicity groups, so that the same mutation in different race groups may have various or even opposite functional effects.
Study of the genetic structure of Ethiopian Jews by using mtDNA and some nonrecombinant Y-chromosome markers and 5’-globin haplotypes elements clearly demonstrated that Ethiopian Jews are a mixture of African and Caucasian (Asian) population and are significantly different from other Jewish communities [42-46]. Several authors argued that Ethiopian Jews derive mostly from Africans [47]; however, both cultural and historic evidence shows tight relationship between populations of Ethiopia and Asia (Near East and southern Arabia).
3.3 Population Divergence
Using the CCR5-CCR2 haplotype frequency estimates, the genetic distances (Corrected Average Pairwise Difference: ARLEQUIN software package) between Ethiopian Jews and other 7 populations were
evaluated (Table 4).
The population relationship reconstructed by using these distances (the UPGMA algorithm: PHYLIP software package) was also undertaken and the results are depicted in Fig. 2.
As can be seen, the dendrogram shows two well-defined population groups: The first one containing two populations, namely Non-Pygmy and American Africans, and the second one containing the remaining six populations, which are further divided. Within these groups, Ethiopian Jews are “sisters” to Asian populations (non-Indians and Indians), while
Thai population is found to be the most distant.
Table 4 Population average pairwise differences*.
Population Non-pygmy Afr. Amer European Non-indian Indian Eth. Jews Thai Argentina
Non-pygmy# 0.78 0.79 0.83 0.85 0.83 0.85 0.87 0.83
Afr. Amer# 0.21 0.79 0.82 0.81 0.82 0.83 0.81 0.81
European# 11.04 7.26 0.66 0.71 0.71 0.72 0.7 0.73
Non-indian# 11.07 7.72 3.14 0.7 0.73 0.71 0.66 0.73
Indian# 7.44 4.55 1.49 1.56 0.73 0.73 0.7 0.75
Eth. Jews 10 6.92 3.64 0.16 0.41 0.72 0.66 0.74
Thai## 19.36 15.17 8.03 2.75 4.91 1.77 0.57 0.7
Argentina# 6.5 3.94 2.45 0.88 0.99 1.09 4.27 0.75
*Above diagonal: Average number of pairwise differences between populations, d(ij). Diagonal elements: Average number of pairwise differences within population, d(i). Below diagonal: Corrected average pairwise difference, d(i, j)-(d(i)+d(j))/2. Row data of CCR5 haplotypes distribution was taken from: # - [32] and ## - [37].
NON-PYGMY
AFR. AMER.
EUROPEAN
NON-INDIAN
ETH. JEWS
INDIAN
ARGENTINA
THAI
Fig. 2 UPGMA tree based on CCR5 haplotype frequencies.
4. Conclusion
No statistically significant differences between HIV-
unexposed and exposed uninfected Ethiopian Jews at
the allele, genotype, or haplotype level have been found.
The pattern of CCR5-CCR2 genetic variations in Eth-
iopian Jews resembles the one found in Asian populations
Acknoledgments
The authors thank M.Malasky for dedicated technical assistance.
References
[1] J.P. Moore, A. Trkola, T. Dragic, Co-receptors for HIV-1 entry, Curr. Opin. Immunol. 9 (4) (1997) 551-562. [2] O.J. Cohen, A. Kinter, A.S. Fauci, Host factors in the
pathogenesis of HIV disease, Immunol. Rev. 159 (1997) 31-48.
[3] E. Gonzalez, M. Bamshad, N. Sato, S. Mummidi, R. Dhanda, G. Catano, et al., Race-specific HIV-1 disease-modifying effects associated with CCR5 haplotypes, Proc. Natl. Acad. Sci. 96 (21) (1999) 12004-12009.
[4] S.J. O’Brien, J.P. Moore, The effect of genetic variation in chemokines and their receptors on HIV transmission and progression to AIDS, Immunol. Rev. 177 (2000) 99-111. [5] J. Tang, B. Shelton, N.J. Makhatadze, Distribution of
chemokine receptor CCR2 and CCR5 genotypes and their relative contribution to human immunodeficiency virus type 1 (HIV-1) seroconversion, early HIV-1 RNA concentration in plasma, and later disease progression, J. Virol. 76 (2) (2002) 662-672.
[6] P.S. Kulkarni, S.T. Butera, A.C. Duerr, Resistance to HIV-1 infection: Lessons learned from studies of highly exposed persistently seronegative (HEPS) individuals, AIDS Rev. 5 (2) (2003) 87-103.
[7] M. Li, R. Song, S. Masciotra, V. Soriano, T.J. Spira, R.B. Lal, et al., Association of CCR5 human haplogroup E with rapid HIV type 1 disease progression, AIDS Res. Hum. Retroviruses 21 (2) (2005) 111-115.
[8] M. Dean, M. Carrington, C. Winkler, G.A. Huttley, M.W. Smith, R. Allikmets, et al., Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene, Science 273 (5283) (1996) 1856-1862.
[9] P.A. Zimmerman, A. Buckler-White, G. Alkhatib, T. Spalding, J. Kubofcik, C. Combadiere, et al., Inherited resistance to HIV-1 conferred by an inactivating mutation in CC chemokine receptor 5: Studies in populations with contrasting clinical phenotypes, defined racial background, and quantified risk, Mol. Med. 3 (1) (1997) 23-36. [10] R. Liu, W.A. Paxton, S. Choe, D. Ceradini, S.R. Martin, R.
Horuk, et al., Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection, Cell 86 (3) (1996) 367-377.
[11] W.A. Paxton, S.R. Martin, D. Tse, T.R. O’Brien, J. Skurnick, N.L. VanDevanter, et al., Relative resistance to
HIV-1 infection of CD4 lymphocytes from persons who remain uninfected despite multiple high-risk sexual exposure, Nature Medicine 2 (4) (1996) 412-417.
[12] M.P. Martin, M. Dean, M.W. Smith, C. Winkler, B. Gerrard, N.L. Michael, et al., Genetic acceleration of AIDS progression by a promoter variant of CCR5, Science 282 (5395) (1998) 1907-1911.
[13] A.M. de Roda Husman, M. Koot, M. Cornelissen, I.P. Keet, M. Brouwer, S.M. Broersen, et al., Association between CCR5 genotype and the clinical course of HIV-1 infection, Ann. Intern. Med. 127 (10) (1997) 882-890. [14] L.G. Kostrikis, Y. Huang, J.P. Moore, S.M. Wolinsky, L.
Zhang, Y. Guo, et al., A chemokine receptor CCR2 allele delays HIV-1 disease progression and is associated with a CCR5 promoter mutation, Nature Medicine 4 (3) (1998) 350-353.
[15] M. Mellado, J.M. Rodriguez-Frade, A.J. Vila-Coro, A.M. de Ana, A.C. Martinez, Chemokine control of HIV-1 infection, Nature 400 (6746) (1999) 723-724.
[16] S. Mummidi, M. Bamshad, S.S. Ahuja, E. Gonzalez, P.M. Feuillet, K. Begum, et al., Evolution of human and non-human primate CC chemokine receptor 5 gene and mRNA: Potential roles for haplotype and mRNA diversity, differential haplotype-specific transcriptional activity, and altered transcrition factor binding to polymorphic nucleotides in the pathogenesis of HIV-1 and simian immunodeficiency virus, J. Biol. Chem. 275 (25) (2000) 18946-18961.
[17] Z. Weisman, A. Kalinkovich, G. Borkow, M. Stein, Z. Greenberg, Z. Bentwich, Infection by different HIV-1 subtypes (B and C) results in a similar immune activation profile despite distinct immune backgrounds, J. Acquir. Immune Defic. Syndr. 21 (2) (1999) 157-163.
[18] M. Carrington, M. Dean, M.P. Martin, S.J. O’Brien, Genetics of HIV-1 infection: Chemokine receptor CCR5 polymorphism and its consequences, Hum. Mol. Genet. 8 (10) (1999) 1939-1945.
[19] M.W. Smith, M. Dean, M. Carrington, C. Winkler, G.A. Huttley, D.A. Lomb, et al., Contrasting genetic influence of CCR2 and CCR5 variants on HIV-1 infection and disease progression, Science 277 (5328) (1997) 959-965. [20] J.H. Abramson, P.M. Gahlinger, Computer Program for
Epidemiologists, PEPI version 3, Brixton Books, London, 1999.
[21] L. Komlos, M. Korostishevsky, I. Halbrecht, D. Vardimon, Z. Ben-Rafael, T. Klein, Possible sex-correlated transmission of maternal class I HLA haplotypes, Eur. J. Immunogenet. 24 (3) (1997) 169-177.
[23] M. Nei, Molecular Evolutionary Genetics, Columbia University Press, New York, 1987.
[24] M. Slatkin, Linkage disequilibrium in growing and stable population, Genetics 137 (4) (1994) 331-336.
[25] M. Slatkin, L. Excoffier, Testing for linkage disequilibrium in genotypic data using the EM algorithm, Heredity 76 (4) (1996) 377-383.
[26] S.W. Guo, E.A. Thompson, Performing the exact test of Hardy-Weinberg proportion for multiple alleles, Biometrics 48 (2) (1992) 361-372.
[27] L. Excoffier, M. Slatkin, Maximum-likelihood estimation of molecular haplotype frequencies in a diploid population, Mol. Biol. Evol. 12 (5) (1995) 921-927.
[28] M. Korostishevsky, I. Kremer, M. Kaganovich, A. Cholostoy, I. Murad, M. Muhaheed, et al., Transmission disequilibrium and haplotype analysis of the G72/G30 locus: suggestive linkage to schizophrenia in Palestinian Arabs living in North of Israel, Am. J. Med. Genet. Part B: Neuropsychiatr. Genet. 141 (1) (2006) 91-95.
[29] PHYLIP Phylogeny Inference Package, Release 3.57c, University of Washington, USA, available online at: http://evolution.genetics.washington.edu/phylip.html, 1995. [30] R. Kantor, J.M. Gershoni, Distribution of the CCR5 gene
32-base pair deletion in Israeli ethnic groups, J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 20 (1) (1999) 81-84.
[31] J.J. Martinson, L. Hong, R. Karanicolas, J.P. Moore, L.G. Kostrikis, Global distribution of the CCR2-64I/CCR5-59653T HIV-1 disease-protective haplotype, AIDS 14 (5) (2000) 483-489.
[32] E. Gonzalez, R. Dhanda, M. Bamshad, S. Mummidi, R. Geevarghese, G. Catano, et al., Global survey of genetic variation in CCR5, RANTES, and MIP-1: Impact on the epidemiology of the HIV-1 pandemic, Proc. Natl. Acad. Sci. USA 98 (9) (2001) 5199-5204.
[33] D.H. McDermott, P.A. Zimmerman, F. Guignard, C.A. Kleeberger, S.F. Leitman, Multicenter AIDS Cohort Study (MACS), CCR5 promoter polymorphism and HIV-1 disease progression, Lancet 352 (9131) (1998) 866-870. [34] S. Mummidi, S. Ahuja, E. Gonzalez, S.A. Anderson, E.N.
Santiago, K.T. Stephan, et al., Geneology of the CCR5 locus and chemokine system gene variants associated with altered rates of HIV-1 disease progression, Nature Medicine 4 (7) (1998) 786-793.
[35] A. Mangano, E. Gonzalez, R. Dhanda, G. Catano, M. Bamshad, A. Bock, et al., Concordance between the CC chemokine receptor 5 genetic determinants that alter risks of transmission and disease progression in children exposed perinatally to human immunodeficiency virus, J. Infect. Dis. 183 (11) (2001) 1574-1585.
[36] S. Louisirirotchanakul, H. Liu, A. Roongpisuthipong, E.E. Nakayama, Y. Takebe, T. Shioda, et al., Genetic analysis
of HIV-1 discordant couples in Thailand: Association of CCR2 64I homozygosity with HIV-1-negative status, J. Acquir. Immune Defic. Syndr. 29 (3) (2002) 314-315. [37] C. Yang, M. Li, K. Limpakarnjanarat, N.L. Young, T.
Hodge, S.T. Butera, et al., Polymorphisms in the CCR5 coding and noncoding regions among HIV type 1-exposed, persistently seronegative female sex-workers from Thailand, AIDS Res. Hum. Retroviruses 19 (8) (2003) 661-665.
[38] P.A. Ramaley, N. French, P. Kaleebu, C. Gilks, J. Whitworth, A.V. Hill, Chemokine-receptor genes and AIDS risk, Nature 417 (6885) (2002) 140.
[39] J.R. Salkowitz, S.E. Bruse, H. Meyerson, H. Valdez, D.E. Mosier, C.V. Harding, et al., CCR5 promoter polymorphism determines macrophage CCR5 density and magnitude of HIV-1 propagation in vitro, Clin. Immunol. 108 (3) (2003) 234-240.
[40] K.R. Fowke, T. Dong, S.L. Rowland-Jones, J. Oyugi, W.J. Rutherford, J. Kimani, et al., HIV type 1 resistance in Kenyan sex workers is not associated with altered cellular susceptibility to HIV type 1 infection or enhanced
β-chemokine production, AIDS Res. Hum. Retroviruses 14 (17) (1998) 1521-1530.
[41] T. Messele, T.F. Rinke de Wit, M. Brouwer, M. Aklilu, T. Birru, A.L. Fontanet, et al., No difference in in vitro
susceptibility to HIV type 1 between high-risk HIV-negative Ethiopian commercial sex workers and low-risk control subjects, AIDS Res. Hum. Retroviruses 17 (5) (2001) 433-441.
[42] A. Zoossmann-Diskin, A. Ticher, I. Hakim, Z. Goldwitch, A. Rubinstein, B. Bonne-Tamir, Genetic affinities of Ethiopian Jews, Isr. J. Med. Sci. 27 (5) (1991) 245-251. [43] U. Ritte, E. Neufeld, M. Broit, D. Shavit, U. Motro, The
differences among Jewish communities-maternal and paternal contributions, J. Mol. Evol. 37 (4) (1993) 435-440.
[44] G. Lucotte, P. Smets, Origins of Falasha Jews studied by haplotypes of the Y chromosome, Hum. Biol. 71 (6) (1999) 989-993.
[45] M.F. Hammer, A.J. Redd, E.T. Wood, M.R. Bonner, H. Jarjanazi, T. Karafet, et al., Jewish and Middle Eastern non-Jewish populations share a common pool of Y-chromosome biallelic haplotypes, Proc. Natl. Acad. Sci. 97 (12) (2000) 6769-6774.
[46] N. Rosenberg, E. Woolf, J.K. Pritchard, T. Schaap, D. Gefel, I. Shpirer, et al., Distinctive genetic signatures in the Libyan Jews, Proc. Natl. Acad. Sci. 98 (3) (2001) 858-863.
First Record of
Frankliniella Occidentalis
and Impatiens
Necrotic Spot Virus in Egypt
Abeer Salah El-Deen Abd El-Wahab, Mohamed Abdel-Kader El-Sheikh and Salah Elnagar
Department of Economic Entomology and Pesticides, Faculty of Agriculture, Cairo University, Cairo 12613, Egypt
Received: December 10, 2010 / Accepted: March 01, 2011 / Published: September 30, 2011.
Abstract: The presence of the western flower thrips Frankliniella occidentalis was detected for the first time in Egypt. This species
was found on ornamental plants as well as weeds grown in the Giza region during two field surveys which were carried out in the flowering seasons (July/August) of 2005 and 2006. Out of the 34 plant species inspected, this thrips was recorded on 27 species. Antirrhinum majus harbored the highest number of the thrips. F. occidentalis was differentiated from the onion thrips Thrips tabaci, which was often encountered on the same plant. The differences between the two species were evident in the number of segments of adult antennae, the pronotum of the prothorax, the adult wings and the 8th abdominal tergite by analyzing mounted specimens. F. occidentalis was encountered on Ipomoea tricolor plants (Convolvulaceae) with prominent signs of a virus infection, from which impatiens necrotic spot virus (INSV) was isolated. The isolate was maintained in a greenhouse on Begonia as a virus source. The virus was sap transmitted to a small range of plants, which developed characteristic symptoms of INSV. Infection was confirmed in all inoculated plants by DAS-ELISA using specific antiserum. Out of the 34 species collected in the field, INSV was found on plants of at least 16 tested plant species. This is the first reported occurrence of INSV in Egypt. Insect transmission tests were carried out using nymphs and adults of either F. occidentalis, F. tritci or T. tabaci species. F. occidentalis proved to be the vector of the virus, while T. tabaci and F. tritici failed to transmit INSV.
Key words: Tospovirus, thrips, Frankliniella occidentalis, impatiens necrotic spot virus (INSV).
1. Introduction
F. occidentalis and Thrips tabaci are known to have
a wide range of plant hosts; over 220 species are recorded for F. occidentalis. Host plant species preferred by F. occidentalis include ornamentals and cut flowers (chrysanthemum, rose, carnation, impatiens, gloxinia, gerbera, aster, primula, cineraria), vegetables like cucumber, lettuce, tomato, beans, bergine, capsicum, and ground-nut, strawberries, and stone fruits [1]. Among the 5,500 thrips species described worldwide only one percent is known as pest species and about ten species have been confirmed as vectors of plant viruses [2].
Tospoviruses are transmitted and spread in nature by
Corresponding author: Salah Elnagar, professor, research
fields: economic entomology and pesticides. E-mail: [email protected].
a limited number of thrips (Thysanoptera: Thripidae) species. F. occidentalis is an efficient vector of the tospoviruses, tomato spotted wilt virus (TSWV) and impatiens necrotic spot virus (INSV) [3], while T. tabaci is an efficient vector of TSWV. These viruses
are mainly acquired by the first larval instar and transmitted by second instars close to pupation and by adults [4]. Since the world-wide expansion of F. occidentalis, INSV became a widespread virus in the ornamental industry of Europe, Japan, the Mediterranean area and the USA [5, 6]. Previously there were no reports of the occurrence of F. occidentalis or INSV in Egypt. We report here the
2
. Material and Methods
Thrips specimens were collected at random in the Giza region from ornamental plants during two field surveys made in the flowering season (July/August) of 2005 and 2006 on the approx. 50 acres university experimental farm. Samples were collected weekly by taking 10 flowers from different plants and dusting them onto white paper. During sampling, thrips from each flower were collected in 70% ethanol until examination by stereo-microscopy.
Thrips specimens were prepared [7] and thrips were temporarily mounted in Canada balsam or Hoyer’s medium such that the body, antennae, wings and legs could be analyzed using the key of taxonomy [8]. Illustrations were made by projecting the specimens to a microscope with a digital camera at magnifications of 10, 40, 100 or 400 times.
Virus-free thrips cultures were produced by confining individual adults of T. tabaci and F. occidentalis on string bean pods. Newly hatched larvae
were further reared and maintained on bean pods [9]. F. tritici was maintained on wheat seedlings. Thrips from
the laboratory culture were reared on infected Begonia source plant in order to provide the viruliferous individuals for the transmission tests.
Virus-infected leaves of Begonia plants showing symptoms of INSV were ground in 0.01 M sodium phosphate buffer, pH 7.0. This extract was inoculated onto indicator plants (Petunia hybrida seedlings) using carborundum as abrasive. The plants were kept under glasshouse conditions, which were natural conditions during July/August; (25 ℃ in average ± 5) for symptom development. Infection on the indicator plants was confirmed by DAS-ELISA using INSV antiserum according to the developed methodology [10]. The antisera were supplied by the Laboratory of Virology, Wageningen Agricultural University, The Netherlands.
For the insect-transmission tests, specimens of thrips species were maintained on potted virus source plants (Begonia seedlings), each species in a separate
compartment. At any transmission test, the tested stage of the thrips (nymphs or adults) were chosen and transferred to healthy indicator plants; either Petunia hybrida seedlings or any tested plant species according
to the experiment. After a 30 minutes inoculation feeding period, the plants were sprayed with an insecticide (Malathion 0.01%). The indicators and test plants were kept in an insect-free greenhouse, routinely sprayed with the insecticide, until the appearance of symptoms.
3. Results
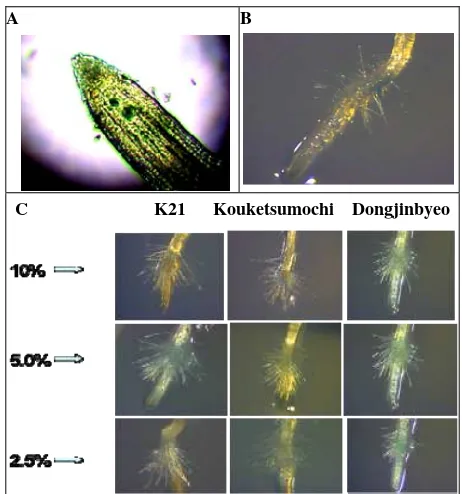
3.1 Identification of the Thrips Collected
T. tabaci could visually be recognized by a larger size and lighter color than F. occidentalis. Their identity was confirmed by weekly mounting at least 10 individuals which were identified by a series of parameters. F. occidentalis species could readily be distinguished from T. tabaci. The antennae of the former consist of eight segments, whereas the latter has antennae with seven antennal segments (Fig. 1A). The pronotum of F. occidentalis has long black setae at the front and back, whereas these setae of T. tabaci are only found at the back (Fig. 1B). Furthermore, the setal structure on the 8th abdominal tergite of F. occidentalis has spines across the abdomen, whereas they are absent
in T. tabaci(Fig. 1C). The flower thrips Frankliniella
tritici (Fitch) is about 1.0 mm long, yellow in color and have pale gray bands or blotches on each segment of the abdomen. It is clearly distinguishable from the other 2 species (Figs. 1A-1C).
3.2 Occurrence of F. Occidentalis and T. Tabaci on Ornamentals
The results of the survey show that F. occidentalis
and T. tabaci occurred on several ornamentals, F.
occidentalis was recorded on 27 host plants of which
Antirrhinum majus harbored the highest number. T. tabaci was recorded on 21 hosts (Table 1).
Fig. 1 A: The antennae of Frankliniella tritici (a) & F. occidentalis (b)have eight segments and those of Thrips tabaci (c)have
seven segments; B: The pronotum of F. tritici has long black setae at the front and the back of the pronotum but incomplete at
the back (a), F. occidentalis has long black setae complete at the front and the back (b), while T. tabaci has them only on the
back (c); C: The setal comb on the 8th abdominal tergite of F. tritici incomplete (a), F.occidentalis carries spines (b), while
Thrips tabaci lacks these spines in the center of this tergite (c). (400× magnification).
that, F. occidentalis proved to be the vector of the virus, while T. tabaci and F. tritici did not transmit the virus.
Three day old larvae of F. occidentalis, born on a virus source plant (infected Begonia) as well as adults emerged from the same source, both transmitted INSV after an inoculation feeding of 30 min on Petunia test plants (29.2% and 52% transmission, respectively) as shown in Table 2. The results of INSV symptoms and plant virus host range are shown in Table 3 and Fig. 2.
4. Discussion
Four thrips species, viz. Thrips tabaci Lindman,
Frankliniella dempfi Priesner, Retithrips aegyptica
Marchal and Heliothrips indicus Bagnal are present in Egypt [11, 12]. The present study shows that F. occidentalis can be added to the list. This species and T.
tabaci are common on various crops and weeds in Giza,
Table 1 Host plants on which the thrips Frankliniella occidentalis, Thrips tabaci and other species were collected in the Giza
area (Frankliniella tritici species were placed in the catergory “Number of other species”).
Plant species No. of thrips
collected
No. of thrips identified
No. of T. tabaci
No. of F. occidentalis
No. of other species
Antirhinum majus 365 20 8 12 0
Begonia tuberhybrida 278 20 5 12 3
Celosia argentea 33 10 2 5 3
Chenopodium album 11 5 5 0 0
C. amaranticolor 24 10 7 3 0
Chrysanthemum morifolium 12 12 5 7 0
Coleus hybridus 16 5 3 2 0
Convolvulus arvensis 180 20 7 11 2
Crinum moorei 45 5 0 5 0
Euphorbia seguieriana 230 20 13 7 0
Gazania rigens 11 11 5 6 0
Gladiolus grandiflora 56 10 3 7 0
Gomphrena globosa 165 15 5 4 6
Hippeastrum hybrid 11 5 0 5 0
H. vittatum 25 10 0 10 0
Ipomoea tricolor 123 10 4 6 0
Kalanchoe blossfeldiana 70 10 2 8 0
Lantana camara 170 10 0 10 0
Pelargonium hortorum 56 10 0 10 0
Petunia hybrida 145 10 4 3 3
Ranunculus ficaria 12 7 0 5 2
Rosa hybrida 89 20 7 13 0
Solanum nigrum 120 10 8 2 0
Tropaeolum majus 17 10 5 5 0
Vicia faba 56 7 3 0 4
Vinca minor 27 5 1 4 0
Zinnia angustifolia 17 10 5 5 0
Table 2 The transmission efficiency of INSV by three days old nymphs and adults of three thrips species tested (10 individuals per test plant for 30 min inoculation feeding period).
Thrips species Adults Larvae
Thrips tabaci *0/20 0/25
Frankliniella occidentalis 13/25 (52%) 7/24 (29.2%) Frankliniella tritici 0/32 0/45
* No. infected plants/No. tested.
on the same plant (Table 1). We also detected a number of specimens of T. tritici on a number of plants. F. occidentalis as well as T. tabaci are major vectors of
tospoviruses. Both species transmit TSWV while INSV is known to be efficiently spread only by F. occidentalis [3, 13]. TSWV is frequently found in ornamental plants as well as in vegetable crops [14]. The present investigation reveals the presence of INSV for the first time in Egypt on several plant species. Out of the three thrips species tested only F. occidentalis proved to be a vector [15].
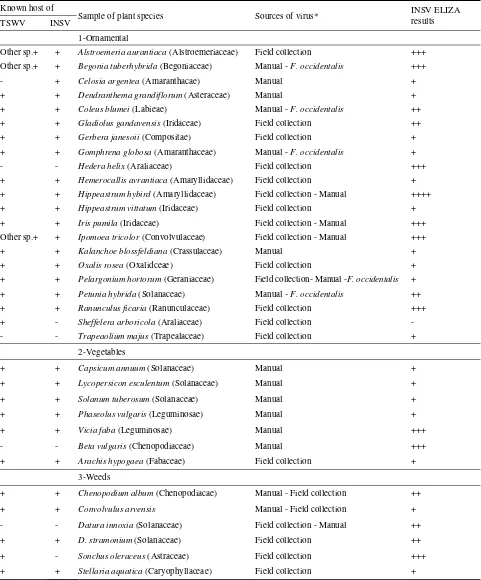
Table 3 Ornamentals suspected to be infected with INSV and the susceptibility of other hosts in Giza area. Known host of
Sample of plant species Sources of virus* INSV ELIZA
results TSWV INSV
1-Ornamental
Other sp.+ + Alstroemeria aurantiaca (Alstroemeriaceae) Field collection +++
Other sp.+ + Begonia tuberhybrida (Begoniaceae) Manual - F. occidentalis +++
- + Celosia argentea (Amaranthacae) Manual +
+ + Dendranthema grandiflorum (Asteraceae) Manual +
+ + Coleus blumei (Labieae) Manual - F. occidentalis ++
+ + Gladiolus gandavensis (Iridaceae) Field collection ++
+ + Gerbera janesoii (Compositae) Field collection +
+ + Gomphrena globosa (Amaranthaceae) Manual - F. occidentalis +
- - Hedera helix (Araliaceae) Field collection +++
+ + Hemerocallis avrantiaca (Amaryllidaceae) Field collection +
+ + Hippeastrum hybird (Amaryllidaceae) Field collection - Manual ++++
+ + Hippeastrum vittatum (Iridaceae) Field collection +
+ + Iris pumila (Iridaceae) Field collection - Manual +++
Other sp.+ + Ipomoea tricolor (Convolvulaceae) Field collection - Manual +++
+ + Kalanchoe blossfeldiana (Crassulaceae) Manual +
+ + Oxalis rosea (Oxalidceae) Field collection +
+ + Pelargonium hortorum (Geraniaceae) Field collection- Manual -F. occidentalis +
+ + Petunia hybrida (Solanaceae) Manual - F. occidentalis ++
+ + Ranunculus ficaria (Ranunculaceae) Field collection +++
+ - Sheffelera arboricola (Araliaceae) Field collection -
- - Trapeaolium majus (Trapealaceae) Field collection +
2-Vegetables
+ + Capsicum annuum (Solanaceae) Manual +
+ + Lycopersicon esculentum (Solanaceae) Manual +
+ + Solanum tuberosum (Solanaceae) Manual +
+ + Phaseolus vulgaris (Leguminosae) Manual +
+ + Vicia faba (Leguminosae) Manual +++
- - Beta vulgaris (Chenopodiaceae) Manual +++
+ + Arachis hypogaea (Fabaceae) Field collection +
3-Weeds
+ + Chenopodium album (Chenopodiacae) Manual - Field collection ++
+ + Convolvulus arvensis Manual - Field collection +
- - Datura innoxia (Solanaceae) Field collection - Manual ++
+ + D. stramonium (Solanaceae) Field collection ++
+ - Sonchus oleraceus (Astraceae) Field collection +++
+ + Stellaria aquatica (Caryophyllaceae) Field collection +
*Only ++ & +++ ELISA results are considered hosts. A single + is usually not enough confirmation. *Field collection = Collected plants from the field showing suspected symptoms.
*Manual = Positive sap test using begonia source plant.
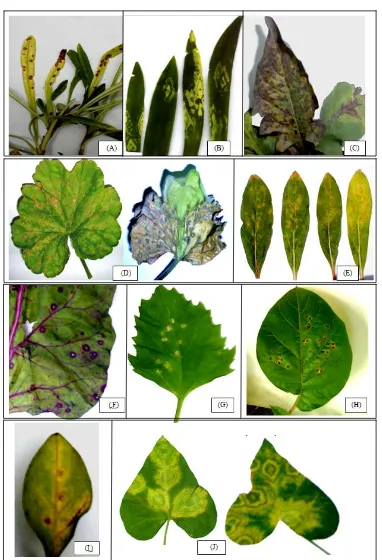
Fig. 2 Symptoms of infection with impatiens necrotic spot virus on several ornamentals in the Giza area. (A): Al stroemeria
aurea; symptoms typical of INSV include pronzed spots, yellow or brown spots or ring spots; (B): Hippeastrum sp. and
Schefflera sp.; the most distinctive symptom of INSV is a thumbprint-like yellow ring spot on leaves; (C): Gerbera sp. showing-
oak leaf symptom. (D): Geranium sp. showing chlorotic developed to necrotic ring spots; (E): Gomphrena globosa showing
small local lesions of INSV not systemic; (F): dark purple ring spots on the leaves of Beta vulgaris; (G): Chenopodium murale,
Ch. album and Ch. amaranticolor, showing chlorotic to necrotic ring spot symptom; (H): Datura innoxia shows symptoms
include black or brown concentric ring patterns on leaves; (I): Petunia hybrida showing local lesions of INSV not systemic; (J):
provide sources and/or reservoirs for virus spread, also cereal weeds; nutsedge (Cyperus esculentus) and purple nutsedge (C. rotundus) in Georgia, are naturally infected with INSV [17]. The relatively high occurrence of INSV found in the sampled fields may explain the recent increase in incidence of INSV in field crops due to the availability of many reservoirs for the virus.
Further studies on the seasonal abundance of thrips species and the impact of INSV on field crops and ornamental plants are in progress.
Acknowledgment
The authors thank Dr. D. Peters, Laboratory of Virology, Wageningen Agricultural University, Binnenhaven, Wageningen 11, 6709 PD, Netherlands for providing the specific antiserum for INSV and for his valuable comments on the manuscript.
References
[1] S.S. Pappu, M.C. Black, H.R. Pappu, T.B. Brenneman, A.K. Culbreath, J.W. Todd, First report of natural infection of peanut (groundnut) by impatiens necrotic spot tospovirus (Family: Bunyaviridae), Plant Dis. 83 (1999) 966.
[2] T. Nagata, D. Peters, An anatomical perspective of tospovirus transmission, in: K.F. Harris, O.P. Smith, J.E. Duffus (Eds.), Virus-Insect-Plant Interaction, Academic Press, San Diego, 2001, pp. 51-67.
[3] I. Wijkamp, N. Almarza, R. Goldbach, D. Peters, Distinct levels of specificity in thrips transmission of tospoviruses, Phytopathology 85 (1995) 1069-1074.
[4] F. van de Wetering, R. Goldbach, D. Peters, Tomato spotted wilt tospovirus ingestion by first instar larvae of Frankliniella occidentalis is a prerequisite for transmission, Phytopathology 86 (1996) 900-905.
[5] M.D. Law, J. Speck, J.W. Moyer, Nucleotide sequence of the 3’ noncoding region and N gene of the S RNA of a serologically distinct tospovirus, J. General Virology 72 (1991) 2597-2601.
[6] M.L. Daughtrey, R.K. Jones, J.W. Moyer, M.E. Daub, J.
Baker, Tospoviruses strike the greenhouse industry: INSV has become a major pathogen on flower crops, Plant Disease 81 (1997) 1220-1230.
[7] J.M. Palmer, L.A. Mound, G.J. Duheaume, Guides to insects of importance to man, in: C.R. Betts (Ed.), Thysanoptera, CAB Int. Inst. Entomol. and British Museum (Natural History), London, 1992, pp. 1-73. [8] L.A. Mound, G.D. Morison, B.R. Pitkin, J.M. Palmer,
Thysanoptera: Key to thrips, requiring carefully prepared microscope slides for examination of minute structures, Handbooks for the Identification of British Insects I (11) (1976) 1-79.
[9] T. Murai, A.J.M. Loomans, Evaluation of an improved method for mass-rearing of thrips and a thrips parasitoid, Entomologia Experimentalis et Applicata 101 (2001) 281-289.
[10] F.M. Clark, N.A. Adams, Characteristics of the micro-plates methods of enzyme linked immunosorbent assay for detection of plant viruses, J. Gen. Virol. 34 (1977) 475-483.
[11] F.C. Willcoks, S. Bahgat, The insects and related pests of Egypt, Sultanic Agric. Soc. 1 (2) (1937) 222-238.
[12] H. Priesner, Genera Thysanopterorum: Keys for the identification of the genera of the order Thysanoptera, Bulletin de la Société Royal Entomologique d’Egypte 33 (1949) 31-157.
[13] L. Tavella, R. Tedeschi, G. Mason, P. Roggero, Efficiency of north-western Italian thrips populations in transmitting tospoviruses, in: R. Marullo, L. Mound (Eds.), Thrips and Tospoviruses, Proceedings of the 7th International Symposium on Thysanoptera, University of Reggio Calabria, 2 July, 2002, pp. 81-86.
[14] EPPO, Impatiens necrotic spot tospovirus, EPPO Bulletin 29 (1999) 473-476.
[15] F.M.J. Assis, S.R. Stavisky, C.M. Reitz, J.L. Sherwood, Mid-gut infection by tomato spotted wilt virus and vector incompetence of Frankliniella tritici, J. Appl. Entomol. 129 (2005) 548-550.
[16] T. Ghotbi, N. Shahraeen, S. Winter, Occurrence of tospoviruses in ornamental and weed species in Markazi and Tehran provinces in Iran, Plant Dis. 89 (2005) 425-429.
Investigation of the Optimum Condition and
Antimicrobial Activities of Pigments from Four Potent
Pigment-Producing Fungal Species
Neveen Saleh Geweely
Department of Botany, Faculty of Science, Cairo University, Giza 12613, Egypt
Received: December 14, 2010 / Accepted: March 01, 2011 / Published: September 30, 2011.
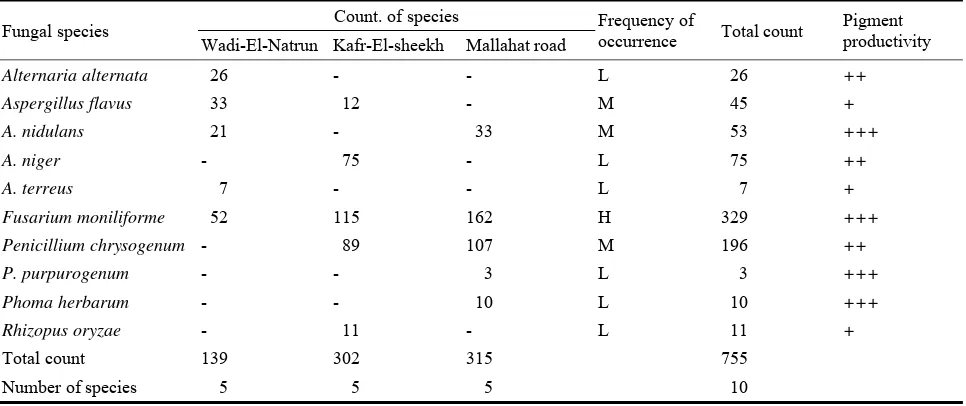
Abstract: Soil samples were collected from three sites (Wadi-El-Natrun, Kafr-El-Sheekh and Mallahat Road) located in Cairo-Alexandria
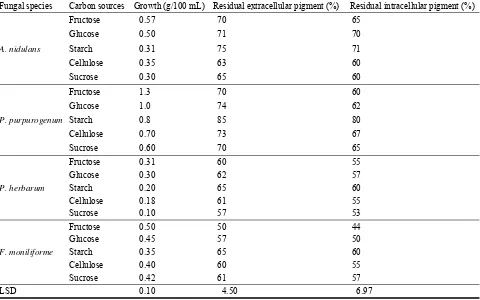
Agriculture Road, Egypt. The total fungal counts allover the road was 755 colonies, constituting ten fungal species (Alternaria alternata, Aspergillus flavus, Aspergillus nidulans, Aspergillus niger, Aspergillus terreus, Fusarium moniliforme, Penicillium chrysogenum, Penicillium purpurogenum, Phoma herbarum and Rhizopus oryzae). The most potent fungal species producing pigments along the road were A. nidulans, F. moniliforme, P. purpurogenum and P. herbarum. Comparative sensitivity to different light wave lengths and radiation (laser, gamma and ultraviolet rays) on growth and pigment production in the four selected fungal species was estimated. Optimization of physical and nutritional factors on growth and pigment production was carried out. A steady increase in the antioxidant activities was showed in all four tested pigments producing species with raising the phenol contents. The extracellular pigment of P. purpurogenum was found to be more effective against some pathogenic microbes and might have a potential role in pharmaceutical drug industry. The identification of the structure of unknown P. purpurogenum pigment was detected using UV and FTIR spectra, and indicated that it is an phenolic compound and has broad stretching OH, C=C and C-H groups of the aromatic ring.
Key words: Fungi, optimum condition, antimicrobial, pigment, antioxidant, phenol.
1. Introduction
Human infections, particularly those involving dermatophytic fungi (Trichophyton, Epidermophyton and Microsporum) and non dermatophytic fungi (Aspergillus, Fusarium and Candida) cause a serious problem, especially in tropical and subtropical developing countries [1, 2]. Dermatophytes are keratinophilic fungi which causing dermatophytosis (tinea or ringworm) in humans and animals [3]. Non-dermatophyte molds are filamentous fungi that are commonly found in nature as soil saprophytes and plant pathogens [4]. Gajera and Vakharia [5] stated that Aspergillus niger is a common contaminant of food. C. albicans caused mucosal and disseminated candidiasis,
Corresponding author: Neveen Saleh Geweely, Ph.D.,
associate professor, research field: microbiology. E-mail: [email protected].