Large-scale sequencing of plant genomes
Steve Rounsley
∗
, Xiaoying Lin and Karen A Ketchum
The large number of ESTs generated forArabidopsisand rice in recent years now act as an important complement to whole genome sequencing projects. TheArabidopsis Genome Initiative has begun a coordinated effort to sequence the entire genome and, as a result, increasing numbers of large sequence entries can be found in the public databases. In addition, the mitochondrial genome ofArabidopsishas been completely sequenced. Genome sequencing studies and the public sequence databases have begun to influence the direction of diverse areas of research from physiology to evolution.
Addresses
The Institute for Genomic Research, 9712 Medical Center Drive, Rockville, MD 20850, USA
∗e-mail: [email protected]
Current Opinion in Plant Biology1998,1:136–141 http://biomednet.com/elecref/1369526600100136
Current Biology Ltd ISSN 1369-5266 Abbreviations
AGI Arabidopsisgene initiative BAC bacterial artificial chromosome EST expressed sequence tag
Introduction
Traditional approaches to isolating genes in plant systems have involved standard molecular and genetic methods, including the screening of expression libraries, homology-based and map-homology-based cloning, and the selection of developmental or physiological mutants. The majority of these experimental strategies are targeted approaches in which a specific gene is the focus of the study and its isolation a prerequisite to further functional characterization. A survey of recent publications in plant molecular biology shows this to be changing. In more and more cases, the starting point for a research project is a DNA sequence found in a public database. Novel genes may be identified in a database search by virtue of sequence similarity with known genes: a bioinformatic process analogous to library screening using nucleic acid hybridization. In yet other cases, interesting genes may be stumbled across by browsing database annotation. Genome initiatives and the Internet resources that contain sequence information have thus dramatically changed experimental plant biology by providing researchers with many more avenues for gene discovery. Here, we review the scope of plant sequencing projects currently underway, the public databases which curate this information and the impact of genomics on plant research.
EST sequencing projects
An expressed sequence tag (EST) is an unedited sequence generated from a single sequencing gel read from a cDNA clone chosen at random from a library. A large-scale EST project can generate partial sequence for a large proportion of genes from a given organism in a very cost-efficient manner. For this reason, such projects have been initiated for many organisms including two key plants,Arabidopsis thaliana and rice (Oryza sativa). There have been two major efforts for Arabidopsis, one by T Newman and colleagues at the Plant Research Laboratory, Michigan State University [2], and another by a French consortium [3]. The two groups proceeded in slightly different directions with the former generating mostly 5′sequences of clones from a single cDNA library constructed using many different plant tissues. In contrast, the French consortium concentrated on generating ESTs from both ends of cDNAs taken from specialized libraries: both tissue-specific libraries and developmental stage-specific libraries were included. The two groups together have generated more than 36,000 ESTs, which have become a major resource forArabidopsismolecular biologists.
In addition to these Arabidopsis projects, there has been a large scale EST project for rice at the Rice Genome Project in Japan which has generated 30,000 ESTs: 20,000 of which are publicly available [4•]. EST sequencing in both projects now continues on a limited basis because of the reduced chances of obtaining novel sequences when such large numbers of ESTs have already been produced. Several commercially funded EST projects are also underway for crop species, particularly maize. Unfortunately these sequence data are not publicly available [5].
rice suggest ∼9,000–10,000 genes are represented by the rice ESTs. A web database available at TIGR has the results of assembling all known transcripts and ESTs into contigs for both rice andArabidopsis. [11–12].
Genomic sequencing
Although EST sequencing projects are very attractive because of the modest investment in cost and time relative to whole genome sequencing, they also have drawbacks. Due to the origin of ESTs, they cannot represent all the genes of an organism and they cannot alone offer insights to gene structure, genomic organization, and evolution. Whole-genome sequencing is an approach that offers the potential to resolve such issues and provides an invaluable resource for future research. The large size of many plant genomes, however, means that this approach is only feasible for very few species. The small size of the A. thaliana genome makes it a logical candidate for whole-genome sequencing and recent publications illustrate several notable achievements towards obtaining the complete sequence.
Sequence data from the nuclear genome of Arabidopsis have been published for several years, usually in the form of a short stretch of sequence spanning a gene of interest. Although important in the study of individual genes, such sequences do not provide many insights into genome organization. The past year has seen an increasing number of large sequence entries in the public databases which span multiple genes. Several individual laboratories have sequenced regions of particular importance to their research (e.g. [13,14•]), whereas data from systematic sequencing by members of the Arabidopsis Genome Initiative (AGI) have begun to flow regularly.
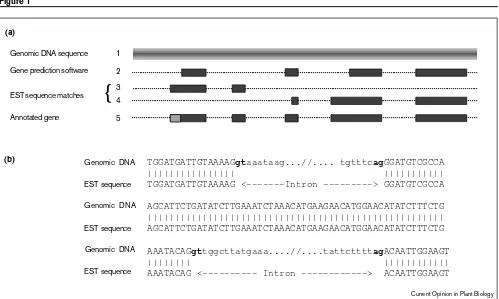
The AGI consists of representatives of each of the sequencing centers participating in the multinational effort to complete the Arabidopsis genome sequence in a rapid and efficient way [15•]. These labs, located in Europe, Japan and the United States, are each sequencing large insert clones on the basis of either bacterial artificial chromosomes (BACs) or bacteriophage genomes (P1 clones) in a systematic manner designed to generate large stretches of contiguous sequence in a defined region of the genome. The sequence of each completed clone is then deposited in the public databases accompanied, in most cases, by annotation describing the genes found within the clone. Annotation of large genomic sequences such as these is distinct from experimentally determined annotation and is achieved by a combination of searching sequence databases and generating predictions of genes with computer software (Figure 1). It is important to realize that accurate annotation depends on ESTs which help define precise intron–exon boundaries. Thus, the EST and whole-genome sequencing projects are complementary.
In addition to the complete sequencing of large insert clones (e.g. BACs), several members of the AGI are
generating end sequences of BAC clones. These short sequence fragments can be used to provide precise overlap information to aid the extension of sequencing contigs and also provide a random sampling of the genome [16]. BAC end sequences, therefore, can be used as additional molecular markers and reveal genes because of the high density of coding regions in theArabidopsisgenome. The BAC end sequences are submitted to the Genome Survey Sequences (GSS) division of GenBank and are searchable at The Institute for Genomic Research (TIGR) and Arabidopsis thalianadatabase (AtDB) websites [17•,18•].
Several recent publications describe initial findings from sequencing portions of the Arabidopsis genome (e.g. [19••,20]). A consortium of European laboratories recently published a description of the sequencing and analysis of a 1.9 Mb region of chromosome 4. Their findings illustrate several features of the genome that are also reflected in the combined AGI sequence. These features include a high gene density which appears to be fairly consistent at around one gene per 5 kb. Of the 389 genes annotated in this 1.9 Mb region, 54% had significant similarity with previously identified genes of known function, whereas 56% matched EST sequences. These numbers are in accord with previously reported estimates based on EST analyses [9•], and with other annotated genomic sequence in the public databases. Another predominant feature described is the presence of clustered gene families indicating a history of tandem duplications (Figure 2). Finally, the localization of retroelements in the genome appears to be as single and double elements, in contrast to the large number of nested retroelements present in maize and other grasses [21,22].
The best source for information regarding AGI progress is the Internet. All annotated genes can be found in the public sequence databases GenBank, European Molecular Biology Laboratory (EMBL), and DNA Database of Japan (DDBJ) which can be conveniently accessed via the services available at the National Center for Biotechnology Information (NCBI) [23••]. These allow searching of the combined sequence information by similarity (BLAST) and by keyword searches (Entrez). In addition, the Ara-bidopsis Database Project at Stanford University (AtDB) [18••] maintains a searchable database of all Arabidopsis sequences (ESTs, BAC ends, and genomic sequences). The progress of all the AGI sequencing projects is also tracked and related back to the physical and genetic maps at AtDB along with links back to the web sites for individual AGI groups [17••,24•,25•–26•,27,28•,31•]. In several cases, these sites display additional data explaining and supporting the annotation provided by the sequencing group.
Impact of plant genome science on research
Figure 1
• • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • •
• • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • •
• • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • • •
TGGATGATTGTAAAAGgtaaataag...//.... tgtttcagGGATGTCGCCA |||||||||||||||| ||||||||||| TGGATGATTGTAAAAG <---Intron ---> GGATGTCGCCA
AGCATTCTGATATCTTGAAATCTAAACATGAAGAACATGGAACATATCTTTCTG |||||||||||||||||||||||||||||||||||||||||||||||||||||| AGCATTCTGATATCTTGAAATCTAAACATGAAGAACATGGAACATATCTTTCTG
AAATACAGgttggcttatgaaa....//....tattcttttagACAATTGGAAGT |||||||| |||||||||||| AAATACAG <--- Intron ---> ACAATTGGAAGT
Genomic DNA
Genomic DNA
Genomic DNA EST sequence EST sequence EST sequence
(a)
(b)
Genomic DNA sequence Gene prediction software EST sequence matches Annotated gene
1 2 3 4 5
Current Opinion in Plant Biology
{
Annotation of genomic sequence using a combination of gene prediction and EST sequence similarity.(a)Graphical representation of the way gene predictions and EST data are used in the annotation of genomic sequence (line 1). Various software programs are available that will predict the location of genes based upon the detection of open reading frames, consensus splice sites, and codon usage from a training set of known genes (line 2). These predictions are not always correct, however, and it is critical to include as much additional information in the annotation process. When available, EST data (lines 3 and 4) very precisely define the location of intron/exon boundaries and improve the annotated gene (line 5) of the transcribed gene. In this example, EST data has also allowed identification of a 5′untranslated region (light gray).
(b)An example of the alignment of genomic and EST sequence data showing the precise identification of intron position that is possible with good quality EST sequences. The consensus GT and AG of splice site donor and acceptor sites are indicated in bold.
These efforts, in conjunction with past and continuing plant EST projects, have had a profound impact on our understanding of gene diversity and the global organization of plant chromosomes. Analyses of plant systems have been further advanced by complementary genome efforts [32••,33••], particularly the sequencing of the mitochondrial genome of Arabidopsis [34••] and the prokaryotic genome of the photosynthetic cyanobacterium Synechocystis [35•]. Highlighted below are several areas where genome science has influenced the direction of plant research.
Multigene families
Although multigene families have been noted inA. thali-ana, for example the AHA gene family that encodes ten isoforms of the plasma membrane proton ATPase [36], the contribution of paralogous sequences was not fully appreciated until large-scale sequencing initiatives were underway. Several reports in 1997 describe extended gene families gleaned from sequence databases [10,37–40]. Duplications have occurred in all functional categories with interesting studies focused on ribosomal proteins [10•], plant defensins [37], transporters [38,39], as well
as biosynthetic enzymes [40]. These data lead one to speculate that multigene families are probably more common than unique coding regions in Arabidopsis, a theme perhaps shared among all multicellular eukaryotes. It seems likely that the abundance of gene families could provide more opportunities for novel gene functions and expression patterns to evolve. The new challenge for plant scientists is to decipher the specific role for each family member in cellular physiology.
Signal transduction
Figure 2
1
10 20 30 40
2 3 4 5 6 7 8 9
9
10
10 2
1 7
3 4 5 6 8 B. napus
Current Opinion in Plant Biology
(a)
(b)
Tandem duplication can lead to large paralogous gene families.(a)The distribution of members of a large gene family similar to a
jasmonate-induced protein fromBrassica napus. The genomic sequence shown here is a 40 kb region from BAC T2O4 (GenBank accession: AC001645). Copy number one is encoded on the opposite strand of DNA to the other nine copies.(b)A comparison of the gene structure for each of the ten copies and theBrassica napusgene. Only one full-length copy exists in this gene cluster (copy number two). Note the variation in the presence of amino-terminal and carboxy-terminal exons and the variation in intron size between members of the family.
abnormalities in stimulus perception or response. The mapping of these mutant alleles to a physically defined region of the genome greatly accelerates map-based cloning strategies for signal transduction factors [42•].
Interactions between nuclear and organellar genomes The completion of theArabidopsismitochondrial genome sequence is a significant step towards understanding the relationship between nuclear and organelle genomes in plants [34•]. Although the Arabidopsis nuclear genome is very compact, it is intriguing that constraints on the size of its mitochondrial genome appear to be fairly relaxed with one coding region per 8 kb. Comparison of stable and variable regions among mitochondrial genomes gives insight into the migration of DNA between eukaryotic compartments. In addition, the large portions of the mitochondrial chromosome with unknown function in Arabidopsis have led to the hypothesis that these intervening sequences may encode guide RNAs necessary forcisandtransRNA editing of the coding sequences.
Communication between nuclear and plastid genomes is also critical in plants. Database searches have helped identify ESTs from the Arabidopsis nuclear genome with homology to eubacterial RNA polymerase sigma factors (sigA, sigB, sigC) [43]. The translated proteins contained
amino-terminal extensions consistent with known plastid-targeting sequences which suggests that the gene products can be imported into the chloroplast. Finally, Northern analysis demonstrated that gene expression for the sig gene family was induced by illumination, supporting the premise that these nuclear-encoded sigma factors may be involved with light-dependent activation of plastid genes [43].
Comparative genomics
The promise of genomics rests on the theory that infor-mation obtained from gene sequences in one organism will have physiological and evolutionary implications for all organisms that maintain a similar coding region. Thus, with each finished genome, evaluation of subsequent chromosomal sequences and their functional relevance have been advanced. This is indeed the case for plant research with information on photosynthesis derived from the model prokaryote Synechocystis [35] and insight into cellular physiology derived from the model eukaryote Sac-charomyces cerevisiae [44••,45•]. Likewise, the Arabidopsis genome will pave the way for comparative analyses of genome sequence from all other plant species.
Conclusion
EST projects in Arabidopsis and in rice have provided partial sequence for a large fraction of the genes from these organisms. In addition, whole-genome sequencing is underway forArabidopsiswhich, in combination with EST data, will provide insights into gene structure, genome organization and the evolution of plants. As a model, the Arabidopsis genome will have a great deal of relevance to all plant species [46].
Arabidopsis will not be the only plant genome that is subjected to such sequencing efforts, however. Similar programs are scheduled for plants with agricultural and economic importance [47]. Funds of $40 million to expand plant genome research have been approved by the United States government and earmarked for the sequencing of cereal crops [48]. Concurrently, a rice genome proposal has received substantial financial support for commencement in Japan [47•]. These parallel programs will benefit greatly from the marked synteny among cereal species [49••] and the policy of international cooperation that has been exemplified by the AGI, the Yeast Genome Project, and the Human Genome Initiative. Genome sequence alone, however, will not furnish all the answers but instead provides a resource that raises many more questions — questions that will occupy plant biologists for decades.
Acknowledgements
We would like to thank Ewen Kirkness and Darrell Doyle for helpful comments on the manuscript. This work was supported by grants from the National Science Foundation, and the United States Department of Energy.
References and recommended reading
Papers of particular interest, published within the annual period of review, have been highlighted as:
• of special interest
•• of outstanding interest
1. Adams M, Kelley JM, Gocayne JD, Dubnick M, Polymeropoulos MH, Xiao H, Merril CR, Wu A, Olde B, Moreno RFet al.: Complementary DNA sequencing: expressed sequence tags and human genome project.Science1991,252:1651-1656. 2. Newman T, de Bruijn FJ, Green P, Keegstra K, Kende H, McIntosh
L, Ohlrogge J, Raikhel N, Somerville S, Thomashow Met al.: Genes galore: a summary of the methods for accessing the results from large-scale partial sequencing of anonymous
ArabidopsiscDNA clones.Plant Physiol1994,106:1241-1255. 3. H ¨ofte H, Desprez T, Amselem J, Chiapello H, Caboche M, Moisan
A, Jourjon MF, Charpenteau JL, Berthomieu P, Guerrier Det al.:An inventory of 1152 expressed sequence tags obtained by partial sequencing of cDNAs fromArabidopsis thaliana.Plant J1993, 4:1051-1061.
•
4. Yamamoto K, Sasaki T:Large-scale EST sequencing in rice. Plant Mol Biol1997,35:135-144.
Summarizes the progress to date in the rice EST sequencing program in Japan.
5. Cohen J:Please pass the data.Science1997,276:1961. 6. Boguski MS, Lowe TMJ, Tolstoshev CM:dbEST — database for
‘expressed sequence tags’.Nat Genet1993,4:332-333.
•
7. dbEST on World Wide Web URL:
http://www.ncbi.nlm.nih.gov/dbEST/index.html
This web site is the central repository for EST sequences from all species. Each EST is regularly searched against the nucleotide and protein databases
and similarity results are stored. Sequence data is also searchable through the BLAST server.
8. Plant Molecular Informatics Center on World Wide Web URL: http://www.cbc.med.umn.edu /ResearchProjects/seq.proc.html
•
9. Rounsley SD, Glodek A, Sutton G, Adams MD, Somerville CR, Venter JC, Kerlavage AR:The construction ofArabidopsis
expressed sequence tag assemblies.Plant Physiol1996, 112:1177-1183.
The process of assembling ESTs into contigs is described and this paper also contains some estimates for a number of genes represented by the EST collection.
•
10. Cooke R, Raynal M, Laudi ´e M, Delseny M:Identification of members of gene families inArabidopsis thalianaby contig construction from partial cDNA sequences: 106 genes encoding 50 cytoplasmic ribosomal proteins.Plant J1997, 11:1127-1140.
Describes a similar approach of assembling ESTs as in [9•] but restricted to the ribosomal proteins.
11. Rice Gene Index on World Wide Web URL: http://www.tigr.org/tdb/ogi/
12. ArabidopsisEST Assemblies on World Wide Web URL: http://www.tigr.org/tdb/at/at.html
13. Tremousaygue D, Bardet M, Dabos P, Regad F, Pelese F, Nazer R, Gander E, Lescure B:Genome DNA sequencing around the EF-1αmultigenic locus ofArabidopsis thalianaindicates a high gene density and a shuffling of non-coding regions. Genome Res1997,7:198-209.
•
14. Quigley F, Dao P, Cottet A, Mache R:Sequence analysis of an 81 kb contig fromArabidopsis thaliana.Nucleic Acids Res
1996,24:4313-4318.
Describes the sequence analysis steps taken for the (at the time) largest contiguous stretch ofArabidopsissequence published. The critical step of sequencing cognate cDNAs was also carried out providing a near perfect gene finding approach. Unfortunately this approach is difficult to scale up.
•
15. Bevan M:Objective: the complete sequence of a plant genome. Plant Cell1997,9:476-478.
Describes the organization of theArabidopsisgenome initiative. 16. Venter JC, Smith H, Hood L:A new strategy for genome
sequencing.Nature1996,381:364-366.
•
17. The Institute for Genome ResearchArabidopsisSequencing Project on World Wide Web URL: http://www.tigr.org/tdb/at/ atgenome/atgenome.html
Provides up-to-date information about the status of sequencing along chro-mosome 2, and a searchable database of all BAC end sequences generated forArabidopsis.
••
18. Arabidopsis thalianaData Base on World Wide Web URL: http://genome-www.stanford.edu/Arabidopsis/
Provides comprehensive access to Arabidopsis information, including progress of the AGI sequencing groups, and searchable databases of all
Arabidopsissequences.
••
19. Bevan M, Bancroft I, Bent E, Love K, Piffanelli P, Goodman H, Dean C, Bergkamp R, Dirkse W, Van Staveren Met al.:Analysis of 1.9 Mb of contiguous sequence from chromosome 4 of
Arabidopsis thaliana. The EU Genome Project.Nature1998, 391:485-488.
Detailed analysis of a substantial portion of theArabidopsisgenome. Fea-tures discussed will probably be typical of most of the genome.
20. Sato S, Kotani H, Nakamura Y, Kaneko T, Asamizu E, Fukami M, Miyajima Net al.:Structural analysis ofArabidopsis thaliana
chromosome 5. I. sequence features of the 1.6 Mb regions covered by twenty physically assigned P1 clones.DNA Res
1997,4:215-230.
21. Voytas DF:Retroelements in genome organization.Science
1996,274:737-738.
22. SanMiguel P, Tikhonov A, Jin Y-K, Motchoulskaia N, Zakharov D, Melake-Berhan A, Springer PS, Edwards KJ, Lee M, Avramova Z, Bennetzen JL:Nested retrotransposons in the intergenic regions of the maize genome.Science1996,274:765-768.
••
23. National Center for Biotechnology Information home page on World Wide Web URL: http://www.ncbi.nlm.nih.gov/
Provides indispensable tools for access to and analysis of sequence data. This site is one of very few that are essential for every biologist to know about.
•
Information about the Stanford group’s progress in chromosome 1 sequenc-ing.
•
25. Genome Sequencing at the ATGC on World Wide Web URL: http://cbil.humgen.upenn.edu/∼atgc/ SPP.html
Information about the ATGC group’s progress in chromosome 1 sequenc-ing, in addition to mapping data and BAC end sequences for clones that hybridize to chromosome 1.
•
26. Plant Gene Expression Center Genome Sequencing Lab on World Wide Web URL: http://pgec-genome.pw. usda.gov/
Information about the PGEC’s role in the SPP sequencing consortium and good general information about sequencing strategies.
•
27. ArabidopsisGenome Analysis at CSHL on World Wide Web URL: http://nucleus.cshl.org/protarab/
Mapping and Sequencing information from chromosomes 4 and 5 for the Cold Spring Harbor Laboratory/ Washington University sequence consor-tium.
•
28. Genome Sequencing Center,Arabidopsispage on World Wide Web URL: http://genome.wustl.edu/gsc /arab/arabidopsis.html Contains a BLAST server of unfinished sequences generated by the CSHL/WU sequencing consortium.
29. CNS Web Site on World Wide Web URL:
http://www.genoscope.cns.fr/externe/arabidopsis /Arabidopsis.html
•
30. KAOS — KazusaArabidopsisdata opening site on World Wide Web URL: http://www.kazusa.or.jp/arabi/
Presents the annotation of the Kazusa sequence from chromosomes 3 and 5, currently the only place the annotation is available.
•
31. MIPSArabidopsis thalianagenome project on World Wide Web URL: http://www.mips.biochem.mpg.de /mips/athaliana/
Contains information about the coordination of the European Scientists’ Se-quencingArabidopsis(ESSA) sequencing project in Europe.
••
32. The Institute for Genome Research database on World Wide Web URL: http://www.tigr.org/tdb/
A collection of curated databases containing DNA and protein sequence, gene expression, cellular role, protein family, and taxonomic data for micro-organisms, plants and humans.
••
33. MAGPIE Automated Genome Project Investigation Environment Database on World Wide Web URL: http://www.mcs.anl.gov /home/gaasterl/genomes.html
Progress reports on sequencing initiatives for virus, phage, eukaryotic or-ganelle, eubacterial, archaeal, and eukaryal genomes. Links to web sites for individual genome projects are provided.
•
34. Unseld M, Marienfeld JR, Brandt P, Brennicke A:The mitochondrial genome ofArabidopsis thalianacontains 57 genes in 366,924 nucleotides.Nat Genet1997,15:57-61. Comparative analysis of theArabidopsismitochondrial genome with mito-chondrial genomes from other species.
•
35. Kaneko T, Sato S, Kotani H, Tanada A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto Set al.:Sequence analysis of the genome of the unicellular cyanobacterium
Synechocystissp. Strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions.DNA Res1996,3:109-136.
First completely sequenced photosynthetic organism, of critical importance to plant biology for functional and evolutionary comparisons.
36. Harper JF, Manney L, Sussman MR:The plasma membrane H(+)-ATPase gene family inArabidopsis: genomic sequence ofAHA10which is expressed primarily in developing seeds. Mol Gen Genet1994,244:572-587.
37. Epple P, Apel K, Bohlmann H:ESTs reveal a multigene family for plant defensins inArabidopsis thaliana.FEBS Lett1997, 400:168-172.
38. Lu Y-P, Li Z-S, Rea PA:AtMRP1gene ofArabidopsisencodes a glutathione S-conjugate pump: isolation and functional definition of a plant ATP-binding cassette transporter gene. Proc Natl Acad Sci USA1997,94:8243-8248.
39. Tommasini R, Vogt E, Schmid J, Fromentau M, Amrhein N, Martinoia E:Differential expression of genes coding for ABC transporters after treatment ofArabidopsis thalianawith xenobiotics.FEBS Lett1997,411:206-210.
40. Pelletier MK, Murrell JR, Shirley BW:Characterization of flavonol synthase and leucoanthocyanidin dioxygenase genes in
Arabidopsis. Further evidence for differential regulation of ‘early’ and ‘late’ genes.Plant Physiol1997,113:1437-1445. 41. Czempinski K, Zimmermann S, Ehrhardt T, Muller-Rober B:
New structure and function in plant K+channels: KCO1, an
outward rectifier with a steep Ca2+dependency.EMBO J1997,
16:2565-2575.
•
42. Mulligan RM, Chory J, Ecker JR:Signaling in plants.Proc Natl Acad Sci USA1997,94:2793-2795.
Timely review of signal transduction research in higher plants stressing po-sitional cloning approaches, and the functional overlap between genes in-volved in the same physiological process.
43. Tanaka K, Tozawa Y, Mochizuki N, Shinozaki K, Nagatani A, Wakasa K, Takahashi H:Characterization of three cDNA species encoding plastid RNA polymerase sigma factors inArabidopsis thaliana: evidence for the sigma factor heterogeneity in higher plant plastids.FEBS Lett1997,413:309-313.
••
44. Goffeau A, Aert R, Agostini-Carbone ML, Ahmed A, Aigle M, Alberghina L, Albermann K, Albers M, Aldea M, Alesandraki D
et al.:The yeast genome directory.Nature1997,387(Suppl):. Describes the landmark achievement of the first completely sequenced eukaryotic genome. Summary of gene content for chromosomes IV, V, VII, IX, XII, XIII, XIV, XV, and XVI and comparisons of the genetic and physical maps.
•
45. Clayton RA, White O, Ketchum KA, Venter JC:The first genome from the third domain of life.Nature1997,387:459-462. Overview of comparative genomics as a tool to define a core set of genes for each domain of life and system-by-system analyses to evaluate functional roles for gene products.
46. Delseny M, Raynal M, Laudi ´e M, Varoquaux F, Comella P, Wu HJ, Cooke R, Grellet F:Sequencing and mapping theArabidopsis
genome: a weed model for real crops.Symp Soc Exp Biol
1996,50:5-9.
•
47. Briggs SP, Helentjaris T:Plant genomics moves into the limelight.Genome Res1997,7:865-857.
Summary of international efforts to fund plant genomic research. 48. “House and Senate approve $40 million for plant genome
initiative” on World Wide Web URL:
http://www.inverizon.com/ ncgi/senspt.htm October1997
••
49. Bennetzen JL, Freeling M:The unified grass genome: synergy in synteny.Genome Res1997,7:301-306.