This work focuses on the regulation of Cdk activity at the end of the cell cycle during mitotic exit. We will demonstrate the ability of the cell cycle motor Cdk to act as its own motor.
Exit from mitosis in Saccharomyces cerevisiae The Mitotic Exit Network (MEN)
Recently, a novel network of proteins has been identified in regulating the release of Cdc14 from the nucleolus during early anaphase (Stegmeier et al., 2002). Cdc5 is thought to act in the MEN pathway through inhibitory phosphorylations of the Bub2/Bfa1 complex, leading to activation of Tem1 (Hu et al., 2001).
Thesis Overview
II describes how phosphorylation of Net1 by Clb-Cdk underlies the disruption of the RENT complex during anaphase and illustrates a fascinating aspect of the switch that governs the return of mitotic cells to the interphase state. Also, given the location of the RENT complex within the nucleolus, interactions of Net1 with RNA Pol I help explain the multifunctionality exhibited by net1-1 mutants.
A multicopy suppressor gene of the G1 cell cycle mutant gene dbf4 of Saccharomyces cerevisiae encodes a protein kinase and is identified as CDC5. Regulation of Cdc28 cyclin-dependent protein kinase activity during the cell cycle of the yeast Saccharomyces cerevisiae.
Summary
Introduction
These observations suggest that Spo12 and Cdc5 cooperate in parallel pathways to remove Cdc14 from Net1 (Visintin et al., 2003). Interestingly, separase- and Cdc5-induced RENT disassembly correlates with Net1 hyperphosphorylation (Shou et al., 2002a; Sullivan and Uhlmann, 2003; Visintin et al., 2003).
Results
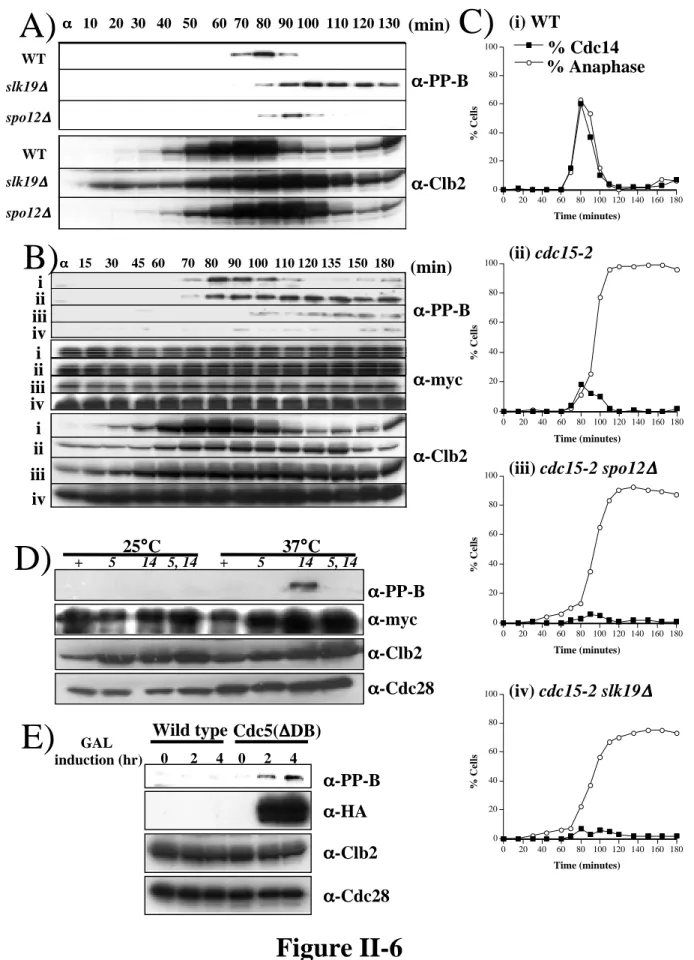
The meiotic defect of net1-6Cdk mutants is less severe than observed in spo12∆, but is very similar to that previously reported for slk19∆ mutants undergoing meiosis ( Buonomo et al., 2003 ). We previously reported that Cdc5 influences the phosphorylation state of Net1 and promotes Cdc14 release in vivo (Shou et al., 2002a).
Discussion
Release of Cdc14 from Net1 during early anaphase requires the FEAR network components Spo12, Slk19, Cdc5 and Esp1 in addition to phosphorylation of Net1 by Clb1,2-Cdk. The FEAR network produces RENT complex disassembly by promoting the phosphorylation of Net1 by Clb-Cdk. Nevertheless, MEN promotes phosphorylation of Net1 by Clb-Cdk, but possibly as an indirect consequence of exposing phosphorylation sites on Net1 that are normally shielded by bound Cdc14.
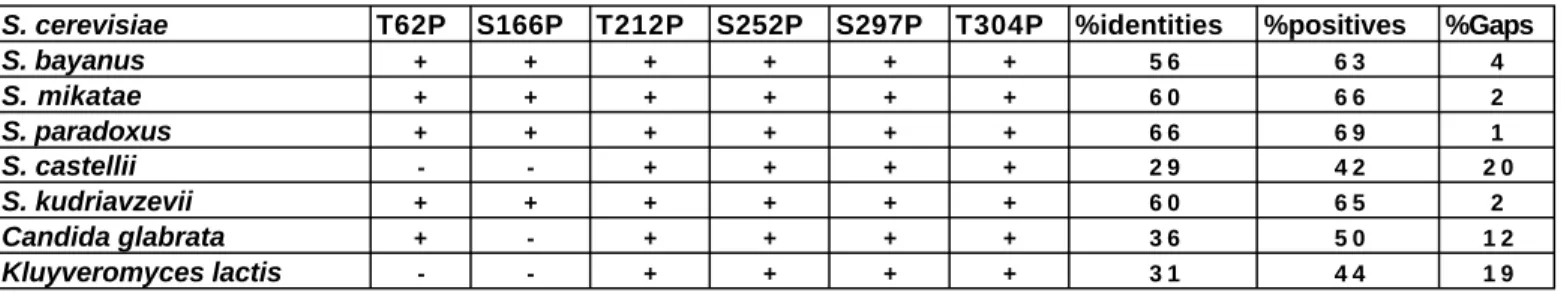
Our data indicate that the FEAR components Spo12, Slk19, and Cdc5 act, at least in part, through phosphorylation of Net1 by Clb-Cdk to promote disassembly of the RENT complex. Furthermore, the early anaphase release of Cdc14 promoted by the FEAR network in a cdc15-2 mutant is transient (Stegmeier et al., 2002), whereas the phosphorylation of Net1 on Threonine 212 is not (Figure 6). Thus, although the FEAR network promotes phosphorylation of Net1 and dissociation of Cdc14 from Net1, the former does not appear to be sufficient for the latter.
In previous work, we reported that Cdc5 affects Net1 phosphorylation and RENT complex disassembly ( Shou et al., 2002a ).
Acknowledgements
Inhibitory phosphorylation of the APC regulator Hct1 is controlled by the kinase Cdc28 and the phosphatase Cdc14. The DBF2 gene product of the Saccharomyces cerevisiae cell cycle is homologous to protein kinases and is periodically expressed during the cell cycle. Inactivation of the cyclin-dependent kinase Cdc28 reverses DNA damage-induced cell cycle arrest and mitotic spindle disassembly in Saccharomyces cerevisiae.
Regulation of the Bfa1p-Bub2p complex at spindle pole bodies by the cell cycle phosphatase Cdc14p. Exit from mitosis is caused by Tem1-dependent release of the protein phosphatase Cdc14 from the nucleolar RENT complex. Destruction of the mitotic kinase CDC28/CLB is not required for the transition from metaphase to anaphase in budding yeast.
The Swi5 transcription factor of Saccharomyces cerevisiae plays a role in mitosis exit by inducing the cdk-inhibitor Sic1 in telophase.
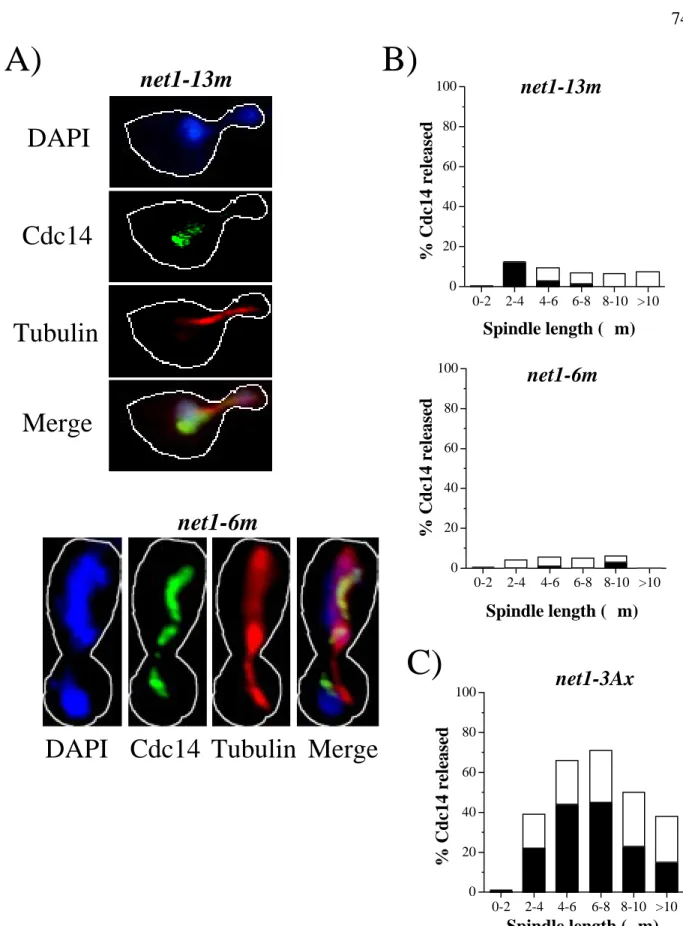
Cdc14 Localization
G1 Anaphase
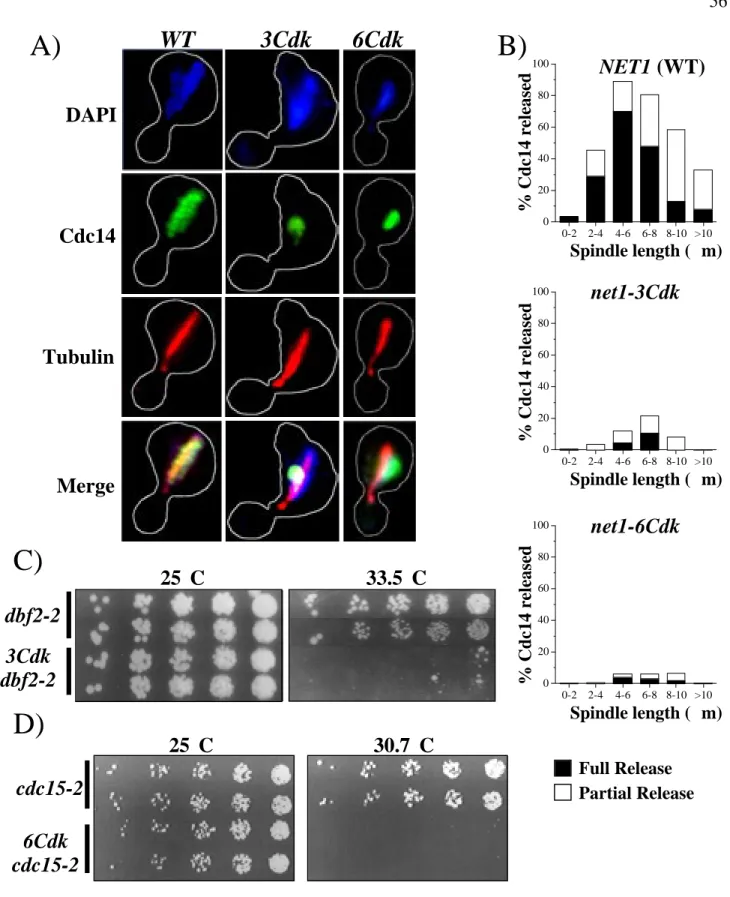
The numbers next to each construct indicate where the transposon insertion occurred in the NET1 locus, with the exception of aa 1-621 of the Net1 HA-fragment, in which the endogenous NET1 was replaced by a truncated allele (RJD1783). Cdc14 localization and microtubule spindle length in asynchronous cell populations were determined by indirect immunofluorescence using anti-Cdc14 and anti-tubulin antibodies, respectively. Hatched lines indicate Net1 fragments with correct nucleolar localization as listed in the TRIPLES database (Ross-Macdonald et al., 1999).
DAPI
Cdc14
Tubulin
Merge
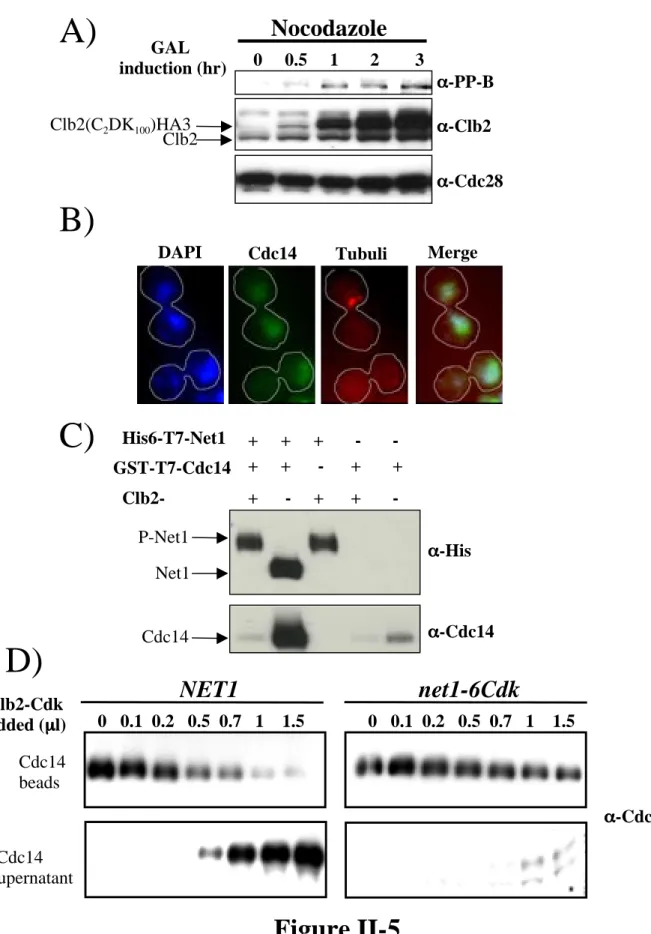
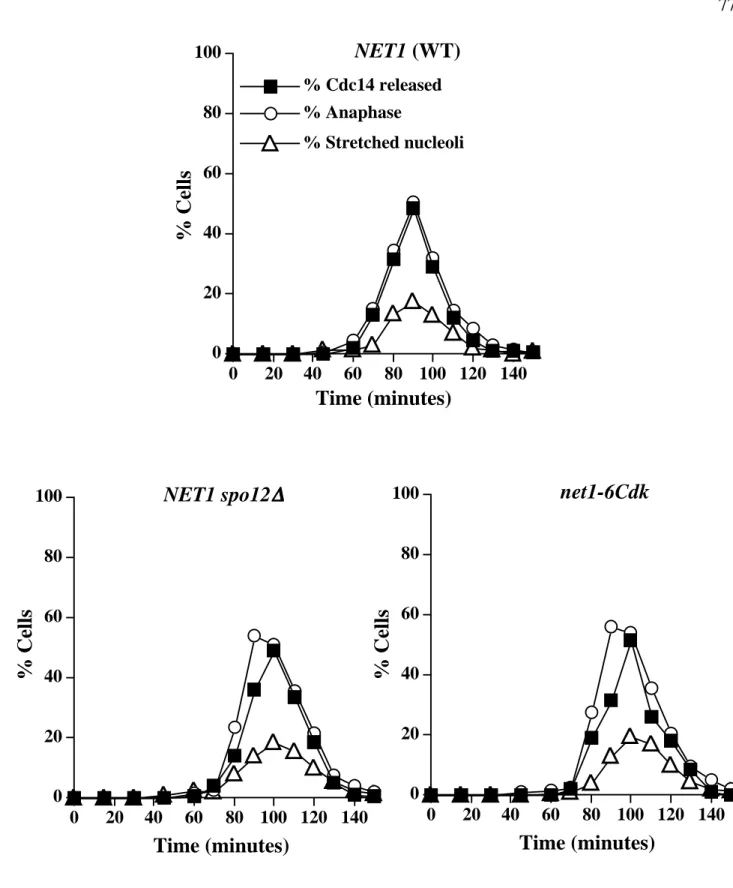
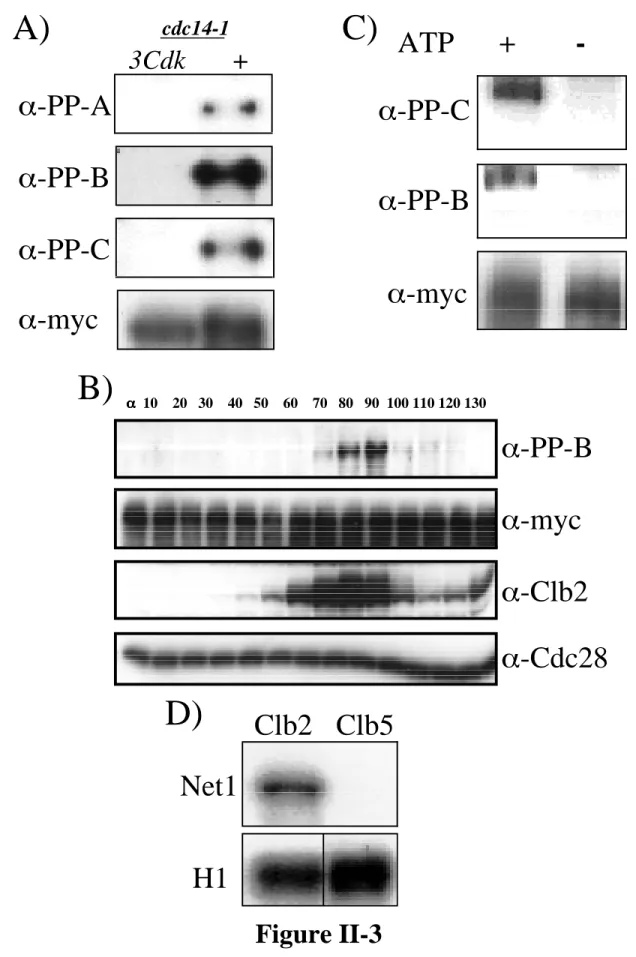
Synchronized cells collected 70 to 110 min after release from α-factor were double labeled with anti-Cdc14 and anti-tubulin antibodies. Anti-myc (9E10) detection of Net1-myc9 was used as a loading control. B) Net1 phosphorylation on Threonine 212 is cell cycle regulated. Cells arrested in α-factor were released from G1 block and samples were collected at the indicated times (min).
Cdc28 levels (α-Cdc28) were used as a loading control. C) Clb2–Cdk generates the in vivo phosphoepitopes in vitro. The bead-bound material was fractionated by SDS-PAGE and immunoblotted with α-PP-B and α-PP-C. Anti-myc (9E10) detection of Net1-myc9 was used as a loading control. D) Net1 is a substrate for Clb2–Cdk in vitro.
Clb2–Cdk and Clb5–Cdk protein kinase complexes were incubated in vitro with Net1 and radiolabeled 32P (upper panel).
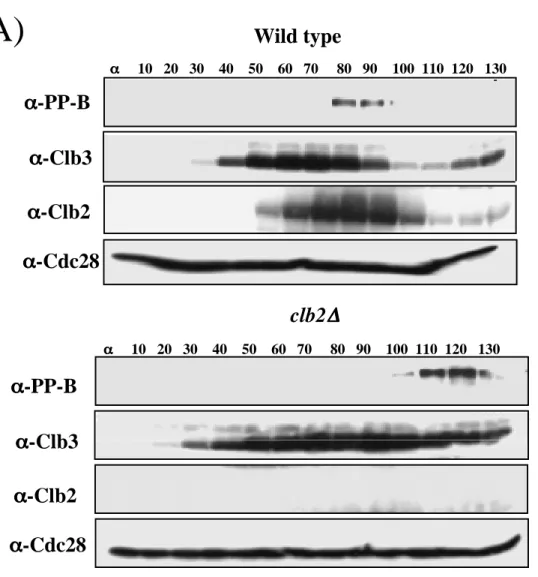
Wild type
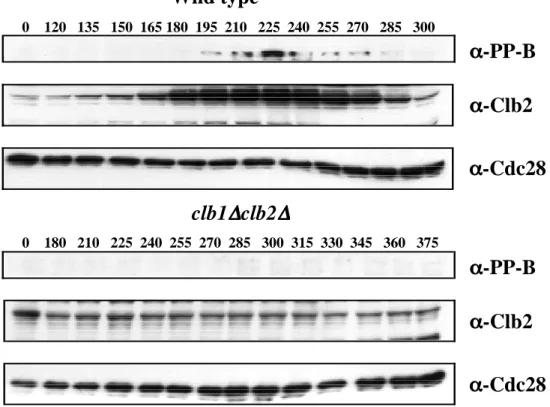
Samples were collected at 10 min intervals, analyzed by SDS-PAGE and immunoblotted with the indicated antibodies. Alpha factor was added back after 70 minutes to prevent cells from entering another cell cycle. Cdc28 levels (α-Cdc28) were used as a loading control. B) Phosphoepitope formation on Net1 is abolished in clb1∆ clb2∆ cells.
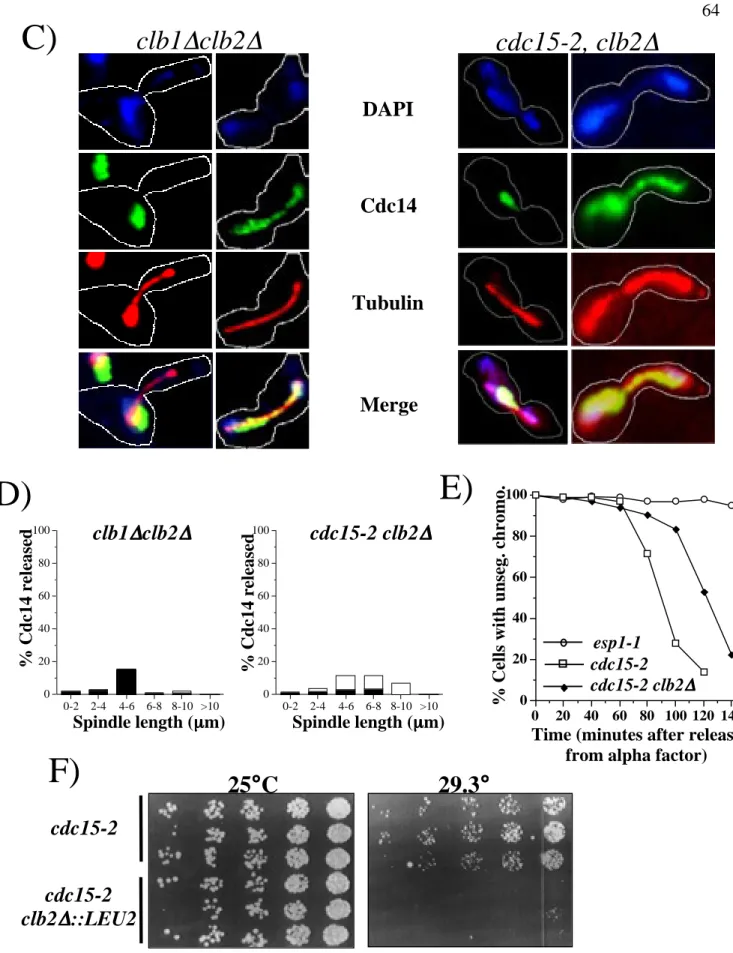
Alpha factor was also added to clb1∆, clb2∆ cells after 300 min in YP 2% glucose to prevent them from entering a second cell cycle. Samples of clb1∆ clb2∆ GAL1p-CLB2 and cdc15-2 clb2∆ cells (RJD 2622) were collected after elutriation/GAL shut-down or α-factor block/release, respectively, for analysis by indirect immunofluorescence with DAPI, anti-Cdc14 and anti-tubulin antibodies. Cells were collected after elutriation and GAL knockdown (clb1∆ clb2∆ GAL1p-CLB2) or α-factor release (clb2∆ cdc15-2) and double labeled with anti-Cdc14 and anti-tubulin antibodies.
Samples were taken at the indicated time points to determine the percentage of cells with unseparated chromosomes (one GFP dot).
Nocodazole
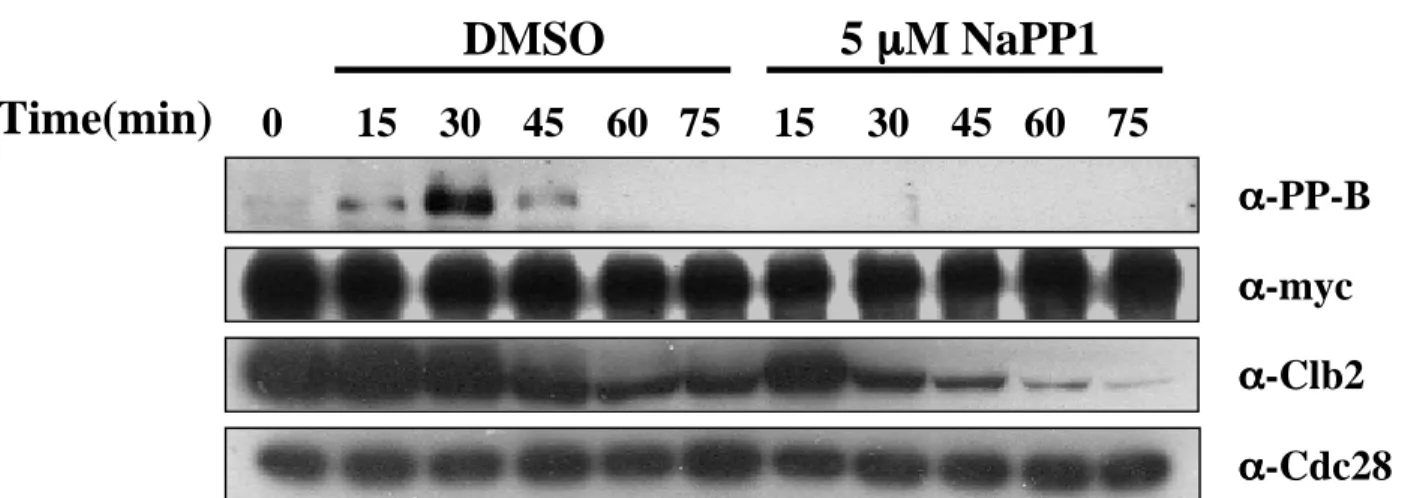
At various times after galactose was added to induce expression of Clb2C2DK100, samples were extracted, fractionated by SDS-PAGE, and immunoblotted with α-PP-B, α-Clb2, and α-Cdc28 as previously described. Staining was performed using DAPI along with anti-Cdc14 and anti-tubulin antibodies as previously described. Cells were released from α-factor block and samples were collected at the indicated time points (min).
The same sections collected for panel (B) were also analyzed by indirect immunofluorescence with anti-Cdc14 and anti-tubulin antibodies, as previously described. At various times after galactose was added to induce expression, samples were withdrawn, separated by SDS-PAGE, and immunoblotted with α-PP-B, α-Clb2, and α-Cdc28 as previously described. Cells harvested at 70 to 110 min after α-factor release were double labeled with anti-Cdc14 and anti-tubulin antibodies.
Cells collected 70 to 110 minutes after α-factor release were double-labeled with anti-Cdc14 and anti-tubulin antibodies.
Time (minutes)
Time (minutes)net1-6Cdk
Aliquots of each culture were collected at the indicated time points for indirect immunofluorescence analysis with anti-Cdc14, anti-RPA190 (nucleolar marker), and anti-tubulin antibodies to monitor Cdc14 release from the nucleolus, nucleolar. The percentage of stretched nuclei was determined by counting cells in which the nucleus had stretched between the mother and daughter cells but had not divided into 2 distinct masses divided by the total number of cells counted.
Time(min)
Transcription of ribosomal DNA repeats is carried out by RNA Polymerase I (Pol I), a holoenzyme composed of 14 proteins (Carles et al., 1991; Huet et al., 1975) and accounts for at least 60% of the total cellular transcript . . Regulation of nucleolar silencing requires a functional RENT complex as well as a functional Pol I as shown by a recent finding that silencing in addition to requiring a functional Sir2 protein also requires transcription by RNA polymerase I (Buck et al., 2002; Shou et al. . , 2001). An intact core is also required for the maintenance of proper silencing as shown by net1-1 and rDNA deletion mutants ( Oakes et al., 1998 ; Shou et al., 2001 ).
Nucleolin, a protein found in the nucleolus of mammalian cells, has been shown to be one of the best substrates for CKII (Caizergues-Ferrer et al., 1987; Schneider and Issinger, 1988). Subcellular localization of CKII has been shown to play an important role in regulation beyond post-translational modification (Faust and Montenarh, 2000; Jans and Hubner, 1996; Tawfic et al., 2001). Furthermore, Net1 has been definitively implicated in regulation of nucleolar structure and Pol I transcription (Shou et al., 2001).
To investigate rDNA segregation in various mutant backgrounds, we chromosomally tagged Net1—which is known to remain in the nucleolus for the entire duration of the cell cycle ( Shou et al., 1999 )—with GFP in various strain backgrounds.
Conclusions
We thank John Keener and the Nomura Laboratory for providing RNA polymerase I antibodies and reagents, Aaron Straight for providing GFP chromosomal constructs, and C.V.C. Division of the nucleolus and its release of CDC14 during anaphase of meiosis I depends on separase, SPO12 and SLK19. Mutational analysis of the structure and localization of the nucleolus in the yeast Saccharomyces cerevisiae.
Temperature-sensitive mutations of the CKA1 gene reveal a role for casein kinase II in the maintenance of cell polarity in Saccharomyces cerevisiae. DNA damage regulation of the RNA components of the translational apparatus: novel biology and mechanisms. The intense staining band found in all lanes around 30kD is TEV protease used for elution of the complex from the beads.
Immunoprecipitation was performed as described in Experimental procedures, and the western blot was immunoblotted with polyclonal anti-Net1 (a gift from D. Moazed) and 9E10 antibodies to detect Net1 and casein kinase II subunits.
DAPI Net1-GFP DIC
Tubulin Cdc14 Merge
Mutant esp1-1, top2-1, and cka2-8 cells containing a lacO array at the LEU2 locus and expressing LacI-GFP from the HIS3 locus were arrested in YP 2% at 37°C glucose. After arrest, samples were taken and analyzed to determine the number of cells with unseparated chromosomes (one GFP point) compared to cells whose chromosomes were separated (two GFP points).
DAPI GFP-LacI DIC
Samples were taken and analyzed by indirect immunofluorescence with DAPI, rabbit anti-Cdc, and rat anti-tubulin monoclonal antibody YL to determine nuclear position, Cdc14 localization, and microtubule spindle length, respectively. The identification of bone kinase substrates for such kinases as Clb-Cdk, Cdc5 and casein kinase II will help us further our understanding of how kinases, by modifying their substrates, play a key role in regulating cell survival and function.
Future Questions