The initial intramolecular electron transfer from high-lying Rydberg states to a covalently bound label with high electron affinity takes place in competition with the Coulomb-stabilized π* orbitals of amide bonds in model peptides. This leads to inhibition of the normal sequence of ECD and ETD processes, which does not result in spine fragmentation.
Background
Selective bond cleavages are often observed (e.g., the C-terminal sides of aspartic/glutamic acids and the N-terminal side of proline12), which is especially useful for de novo sequencing. Ultimately, the amide bonds in the charge-reduced ion are cleaved, leading to the formation of c- and z-type ions.
Contents of Thesis
Investigation of Ion Activation Methods
To investigate their mechanisms, model peptides with a deuterated β-carbon at the disulfide bond are used. Density functional theory calculations are performed to explore the energetics of the proposed disulfide bond cleavage mechanisms.
Protein Quantification and Structural Studies
In the last chapter, we present novel clickable cross-linkers (CXLs) for the sensitive and selective detection of cross-linked peptides from complex mixtures. CXLs are amine-reactive homobifunctional cross-linkers consisting of a central tertiary amine linked to a terminal alkyne.
Conclusion
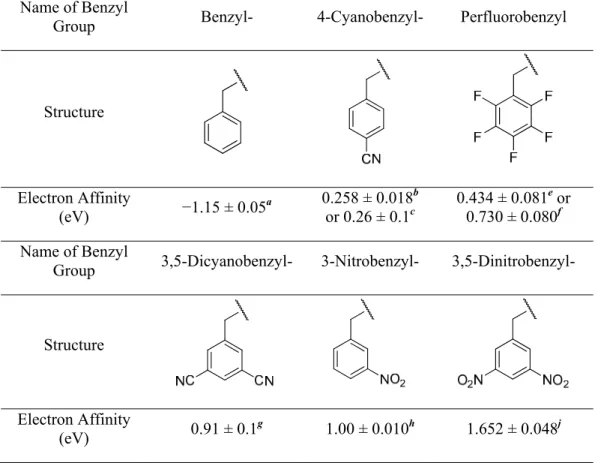
Investigate the mechanism of electron capture and electron transfer dissociation using variable electron affinity labels Transfer dissociation using variable electron affinity labels. Abstract Electron capture dissociation (ECD) and electron transfer dissociation (ETD) of doubly protonated electron affinity (EA) tuned peptides were studied to further elucidate the mechanism of these processes.
Introduction
The derivatized peptide indications generated by electrospray are analyzed by ECD and ETD to investigate the effect of the EA tuning tags. We find that, at sufficiently high EA, the tag leads to inhibition of the spine dissociation process normally observed in ECD and ETD experiments.
Experimental Section
- Materials
- Synthesis of the EA-Tuning Tags and Derivatized Peptides
- Mass Spectrometry
- Quantum Mechanical Calculation
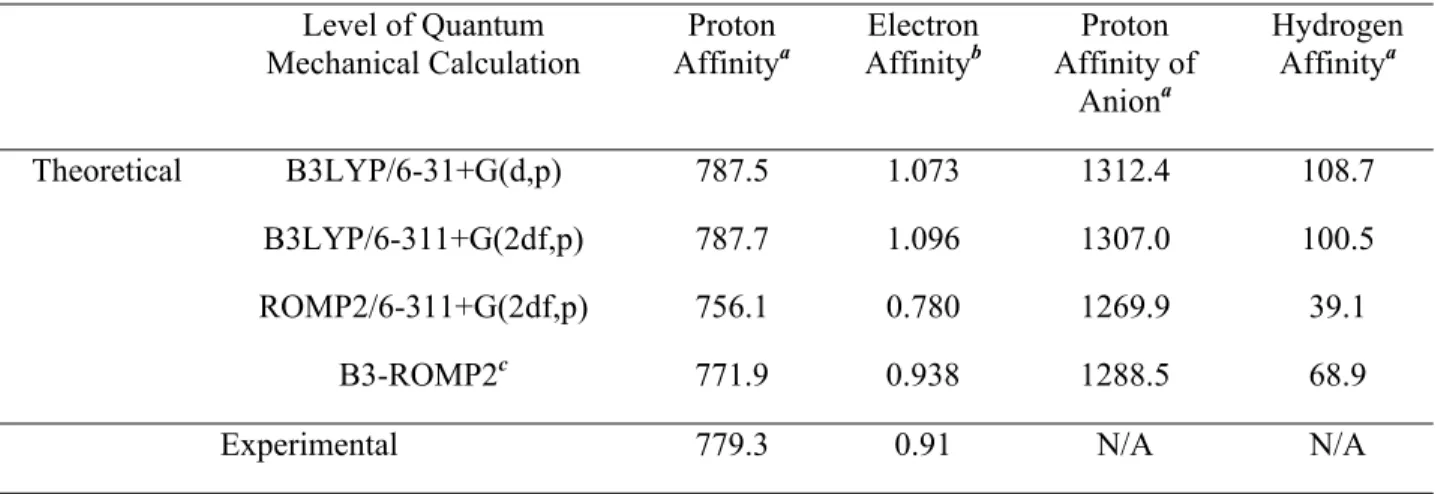
The M06 density functional131 with the same basis sets was also used to estimate the energy of the electron capture process. Calculations of the energetics of excited-state vertical electron capture were performed using time-dependent density functional theory (TDDFT) at the UB3LYP/6-31++G(d,p) and 6-311++G(2df,p) level as implemented in GAMESS for open-shell systems.
Results
ECD of the EA-tuned Peptides
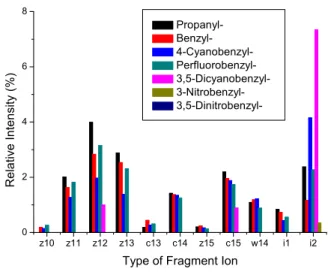
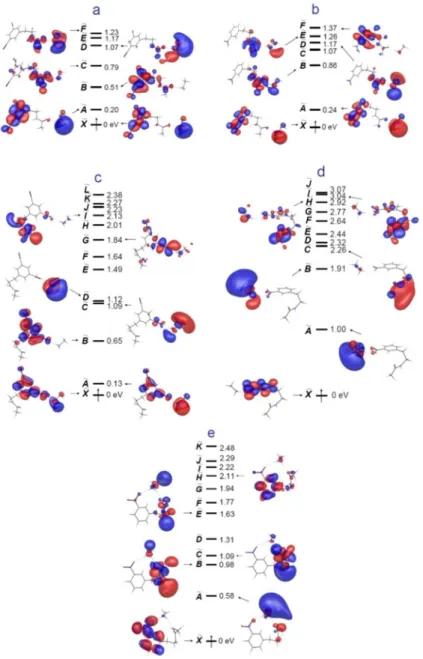
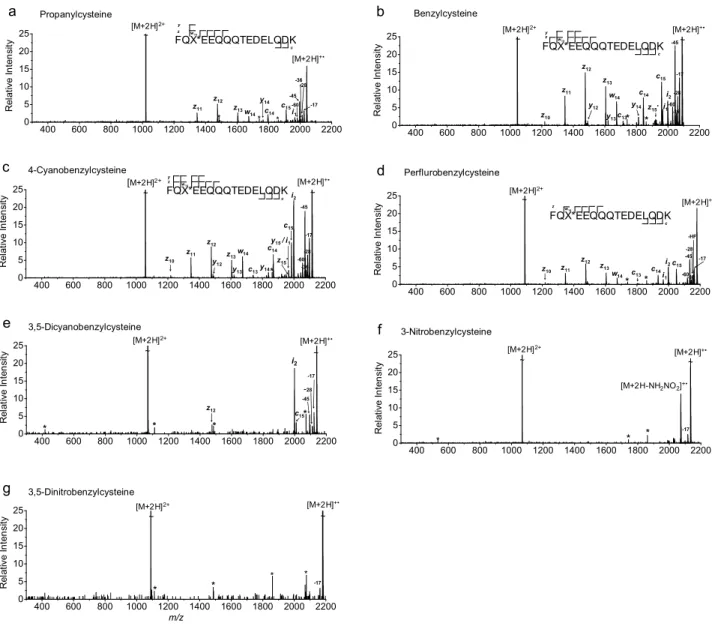
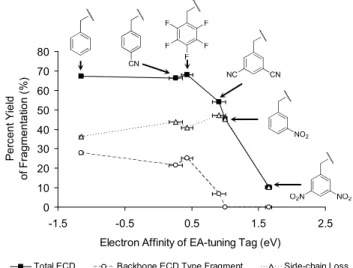
As the EA of the tag increases, the relative abundance of ECD-type ions decreases (Figure 2.1). The ECD spectrum of doubly protonated Na-3,5-dinitrophenyl-derivatized peptide was also examined to demonstrate the effect of the position of the 3,5-dinitrophenyl group and its connectivity (from thioether to secondary amine) in the model peptide .
IRMPD/ECD of the EA-tuned Peptides
Introduction
In Section 3, we extend our FRIPS method to the analysis of disulfide bonds in peptides. The use of CXL in more complex systems (eg in vivo cross-linking) was also tested using HEK293 cells, showing good cell permeability and water solubility.
Probing the Mechanism of Electron Capture and Electron Transfer Dissociation Using
ETD of the EA-tuned Peptides
No evidence for the presence of [b+1]+• and [y+1]+• ions was found in an examination of isotope distributions of b and y ions. However, IRMPD/ECD of [35DCB+2H]2+ exhibits slightly more abundant fragment yields compared to the corresponding ECD and ETcaD spectra (Figures 2.1e, 2.3e and 2.4e).
Hydroxyl Radical Loss and Ion Formation Mechanism in MALDI Plumes
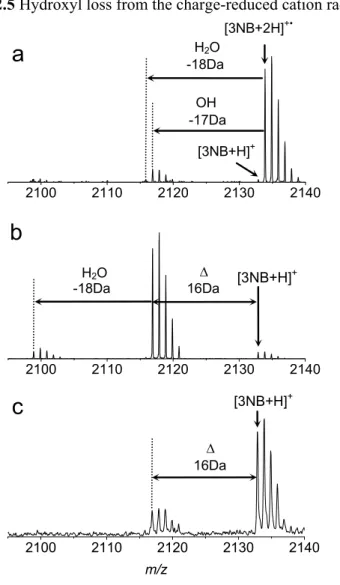
This also indicates that the nascent [35DCB+2H]+• cation radical is less stable compared to [3NB+2H]+• . and [35DNB+2H]+• under the higher level of vibrational excitation. Hydroxyl loss from the charge-reduced cation radical of 3-nitrobenzylcysteine-containing peptide. a) ECD, b) IRMPD/ECD and c) MALDI-TOF MS of 3-nitrobenzylcysteine-containing peptide.
Kinetics of Electron Capture
Variation of the natural logarithm of [B+2H]2+ and [3NB+2H]2+ with electron irradiation time in the ICR cell. The slopes indicate that the electron capture rate is unaffected by electron predation.
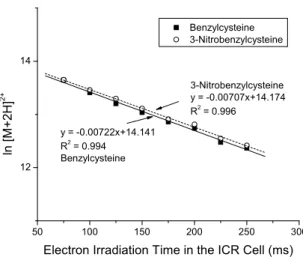
Discussion
- Effect of EA-tuning Tags on Nascent Cation Radicals
- Quantum Mechanical Calculations
- Comparison of ECD, ETD and the Effect of Augmented Vibrational Excitation 48
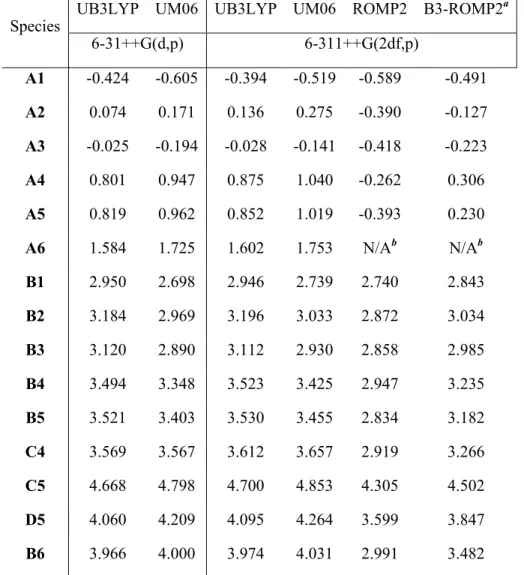
Vertical electron affinities and vertical recombination energies of the model compounds described in Figure 2.8 at different levels of theory. The two methods also have different recombination energies modified by the EA of the electron transfer reagent.
Acknowledgement
Investigation of the mechanisms of inter- and intramolecular disulfide bond cleavages in model peptides by covalent disulfide bond cleavages in model peptides by covalently attached acetyl radical. Abstract We investigate the mechanism of disulfide bond cleavage in gaseous peptide ions induced by a regiospecific covalently attached acetyl radical.
Introduction
In this paper, we use our FRIPS methodology to characterize disulfide bond connectivity in peptides and investigate their disulfide bond cleavage mechanisms. Acetylation of the model peptide AARAAACAA followed by disulfide bond formation with the parent peptide provides a structural mimic (5) of the regiospecific acetyl radical peptide cation (3) generated by FRIPS reagent conjugates (1 or 2).
Experimental
- Peptide Fragmentation Nomenclature
- Materials
- Synthesis of TEMPO-based FRIPS Reagent
- Mass Spectrometry
- Quantum Chemical Calculation
We show that activation of both species leads primarily to cleavage of the disulfide bond. See Supporting Information for details on the synthesis of the TEMPO-based FRIPS reagent and peptide conjugation.
Results and Discussion
- VIP (1-12)
- Arg8-Vasopressin
- Arg8-Conopressin G
- AARAAACAA Dimer
- Deuterium-Labeled AARAAACAA Dimer
- Quantum Chemical Computations
Investigation of the Mechanisms of Inter- and Intramolecular Disulfide Bond Cleavages
Disulfide Bond Cleavage by FRIPS versus ECD
As we discussed in the previous sections, the reaction mechanisms of FRIPS and ECD leading to disulfide bond cleavage are different. The difference in relative abundances of the products of the C−S bond cleavage in each method can be partially attributed to the time scale of measurement.
Reactivity of Disulfide Bond with Radicals
However, the observed disulfide bond cleavage products using the FRIPS and ECD methods share a common feature in that they have visible S−S bond cleavage products. Consequently, preferential S−S bond cleavage fragments are seen in ECD spectra, while competing C−S bond cleavage processes are observed in FRIPS spectra.
Conclusion
Acknowledgment
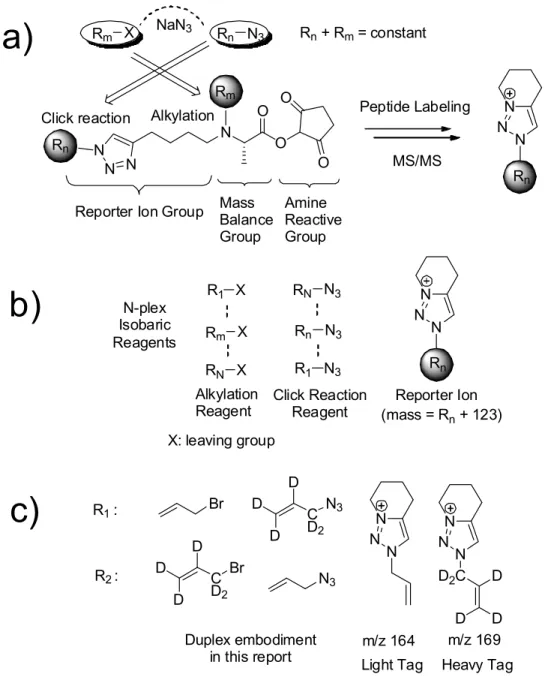
Here, we develop new isobaric tags for protein quantification, called Caltech isobaric tags (CIT), which offer several advantages over other isobaric tags (eg, iTRAQ and TMT). These include 1) generation of reporter ions based on a newly discovered low-energy fragmentation pathway, a nucleophilic displacement reaction with a 1,2,3-triazole ring, 2) in principle an unlimited number of isobaric combinations of CIT reagents, 3) easily adjustable reporter ion mass to access clear windows of m/z values that do not overlap with peptide MS/MS fragments and to avoid low mass exclusion problems typical of ion trap mass spectrometers, and 4) a synthetic methodology that allows the preparation of CIT with minimal costs and effort. These advantages are demonstrated by the preparation of duplex CIT reagents whose reporter ions appear at m/z 164 and 169.
Introduction
After applying a perturbation to one of the light or heavy cell populations, the two samples are pooled for MS analysis. Direct comparison of peptide signals from light- and dark-labeled cell populations yields relative protein expression levels.
Experimental Sections
- Materials
- Synthesis of CITs
- Synthesis of iTRAQ-113 Reagent
- Protein Mixture Digestion
- Affinity Purification and Digestion of Cul1 and Its Associated Proteins
- CIT Labeling
- Instruments
- Data Processing
- Density Functional Calculation
After consumption of the starting material, the mixture was cooled to room temperature, diluted with diethyl ether and filtered. After complete consumption of the starting material by monitoring TLC, the reaction mixture was used for peptide labeling without further purification.
Results and Discussion
- Rationale of CIT Design
- MS/MS of CIT-Labeled Peptides
- Chromatographic Separation
- Protein Labeling
The formation of the reporter ion is simulated by the N-protonated N,N-dimethyl-4-(1-methyl-1H-1,2,3-triazol-4-yl)butan-1-amine. PQD of the 1:1 mixture of light and heavy CIT-labeled peptides in LTQ-Orbitrap produces abundant reporter ions and sequence ions (Figures 4.6a-b).
Conclusion
Acknowledgement
Designer Reagents for Mass Spectrometry-Based Proteomics: Click Chemistry Facilitates
Introduction
Recently, mass spectrometry (MS)-based analysis has enabled detection of binding partners and specific contact residues in more sensitive ways. In vitro cross-linking and enzymatic digestion produce cross-linked peptides that contain spatial information between residues that interact with the cross-linker. For selective and sensitive detection of cross-linked peptides, functionalized chemical cross-linking reagents are required.
Experimental Section
- Materials
- Synthesis of Clickable Cross-Linker (CXL)
- Cross-Linking of a Model Peptide
- Cross-Linking of Ubiquitin
- In vivo Cross-Linking of Cul1
- Mass Spectrometry
- Circular Dichroism Spectrometry
- xQuest Search
- X-ray Crystal Structure Analysis
Forty µg of the cross-linked tryptic digest of ubiquitin was subjected to click reaction by combining resulting peptides from two identical cross-linking experiments. The CXL-crosslinked Ac-AAKAAAAAKAR model peptide was analyzed with an LCQ-deca XP ion trap mass spectrometer (Thermo Fisher Scientific, San Jose, CA).
Results and Discussion
- Model Peptide Cross-Linking
- Structural Analysis by Circular Dichroism
- Ubiquitin Cross-Linking
- CID and ETD of Cross-Linked Peptides
- Sample Clean-up Following Click Reaction
- Peptide Fractionation by SCX
- Avidin Affinity Chromatography
- Validation of Cross-Linked Residues
- In vivo Cross-Linking of HEK 293
- Application for Complex Systems
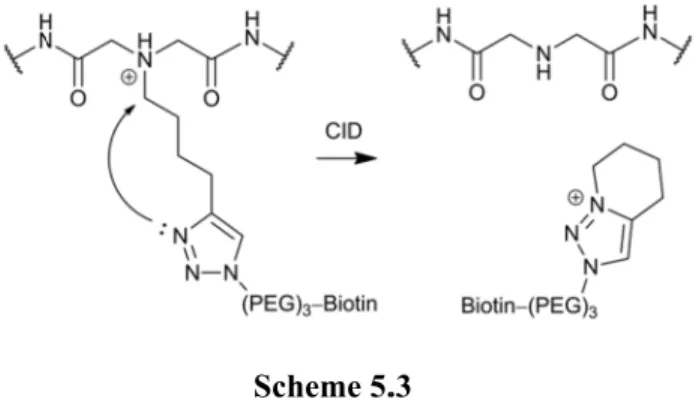
As summarized in Tables 5.1 and 5.2, highly charged cross-linked peptides elute in the high concentration range. Here, monomeric avidin affinity chromatography was used to enrich cross-linked peptides from a simple ubiquitin-cross-linked sample prepared without SCX fractionation (Figure 5.4c).
Conclusions
Designer Reagents for Mass Spectrometry-Based Proteomics: Clickable Cross-Linkers
In CID, peptide fragmentation patterns are highly dependent on the sequence and charge state of the peptide ion. Despite the many efforts of the last decade, there is still much to learn about the mechanistic details of ZHF and ETD.
All vertical electron affinities and recombination energies of the model systems were calculated without geometric relaxation. In addition to Figure 2.1a, the spectra are presented in order of increasing EA of the benzyl substituents.
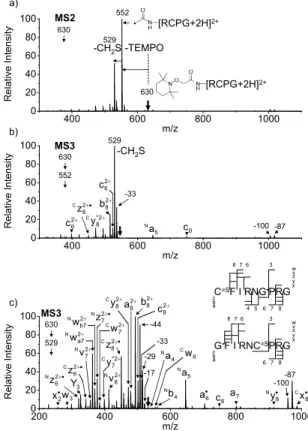
Collisive activation of the acetyl radical cation (m/z 1125) mainly induces CH2S loss (m/z 1079) by cleaving the S−S bond. Collisive activation of the acetyl radical dication (m/z 552) mainly produces the cleavage product of the disulfide bond (m/z 529) via CH2S loss (Scheme 3.5).
The first step of this pathway, H-abstraction reaction at the α-carbon, is energetically favored over abstraction at the β-carbon (tertiary versus secondary hydrogens). We will later explore this mechanism with intermolecular disulfide bond containing peptides that may experience less steric hindrance to H abstraction at the α-carbon via the remote mode rather than the direct 1,4 interaction.
The regioselective acetyl radical group is generated by collision activation of the derivatized AARAAACAA peptide dimer with the TEMPO-based doubly protonated FRIPS reagent (2HH, m/z 873). In addition, the energy of direct substitution of the acetyl radical on the sulfur atom, followed by cleavage of the S−S bond, is considered (Figure 3.10).
The design of new reagents is hindered by the fact that very few low-energy fragmentation pathways suitable for the production of the reporter ions are known in peptide tandem MS.
CIT-labeled peptides are fragmented by various ion activation methods (e.g. PQD, beam-type CID, and High Energy Collisional Dissociation (HCD)), yielding the reporter ions whose mass is Rn + 123 Da. c) the duplex version of the CIT reagents in this report using allyl bromide-d0 and d5. MALDI TOF MS spectra of a) light and b) heavy CIT reagent labeling of the model peptide, VIP (1-12), HSDAVFTDNYTR.
Due to the low complexity of the ubiquitin cross-linked sample, SCX fractionation by itself is sufficient for separation and identification of cross-linked peptides from other linear peptides (Figure 5.4d-g, Table 5.1 and 5.2). Using chemical reactions and subsequent sample treatments, less abundant cross-linked peptides can be lost (Table 5.2).