Through cell analysis, various tasks such as cancer diagnosis, reconstruction of synaptic connectivity maps, measurement of drug response and so on could be possible. With the advent of recent advances in deep learning, more accurate and high-throughput cell segmentation has become possible. However, deep learning-based cell segmentation faces a problem of cost and scalability to construct datasets.
In this thesis, Scribble2Label, a new weakly supervised cell segmentation framework that exploits only a handful of scribble annotations without full segmentation labels. For this, we exploit the consistency of predictions by iteratively averaging the predictions to improve pseudo-labels. The performance of Scribble2Label is demonstrated by comparing it to several state-of-the-art cell segmentation methods with various cell imaging modalities, including bright field, fluorescence, and electron microscopy.
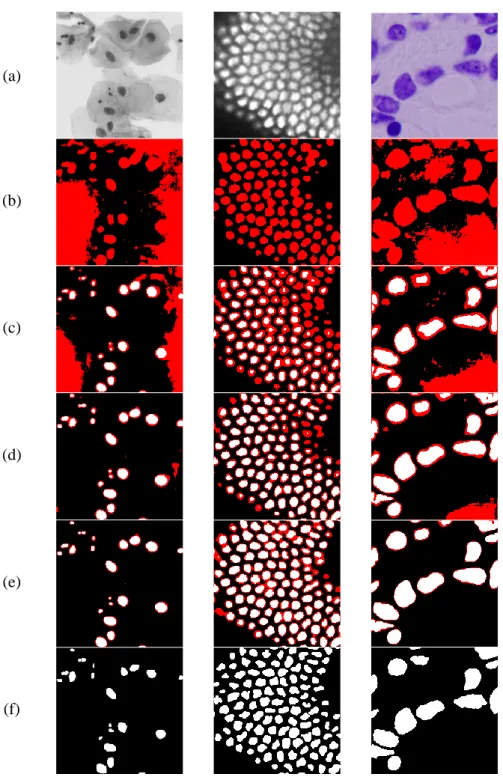
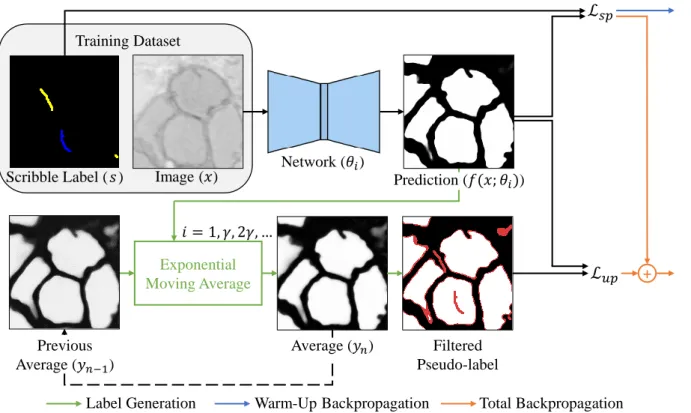
Then Lsp is calculated with the scribble annotation, and Lupis is calculated with the filtered pseudolabel. The red is the pixel below the consistency thresholdτ, the white and black is the cell or background pixel above τ. a) is an input image, (b)-(e) are the label generation results as training progresses, (f) is a complete label. We can observe that the self-generated label gets close to the ground truth label as pseudo label is refined.
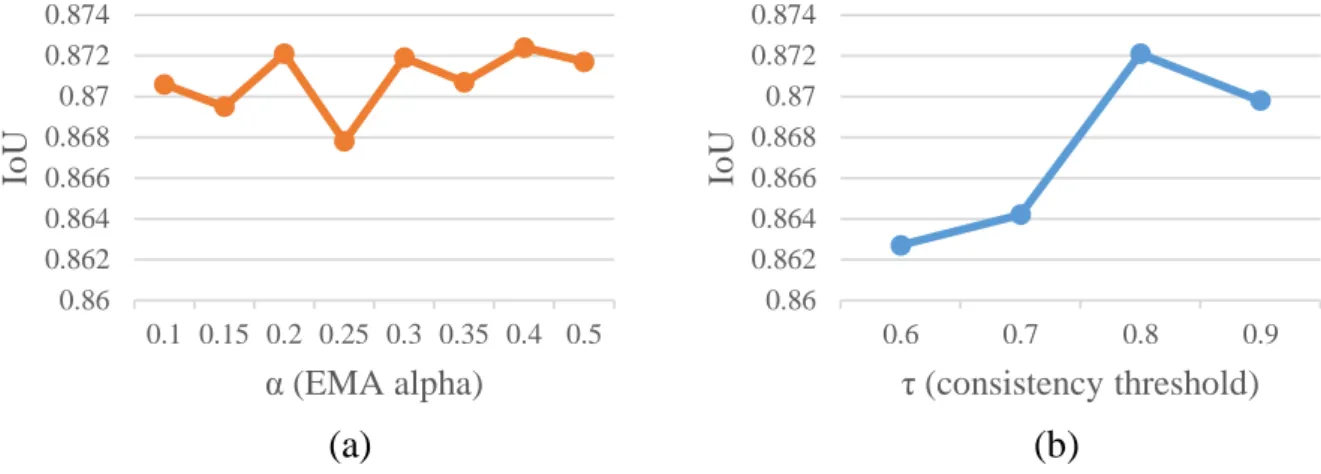
Varying the consistency threshold to measure whether the generated label of the scratchable region is reliable using prediction ensemble.
Problem Definition
In addition, manually generated segmentation labels are prone to errors due to the difficulty of drawing area masks at the pixel level. To solve such problems, poorly supervised cell segmentation methods using point labels have recently been proposed [12–14]. Although generating point annotations is much easier compared to full regional masks, existing work requires point annotations for the entire dataset—for example, there are around 22,000 nuclei in 30 images of the MoNuSeg dataset [ 4 ].
Moreover, the performance of the work mentioned above is highly sensitive to the point location, i.e. the point must be close to the center of the cell. Recently, weak-supervised learning using scribble annotations, that is, scribble-supervised learning, has been actively studied in image segmentation as a promising direction to reduce the burden of manually generating training labels. Scribble supervised learning exploits scribble labels and regularized networks with standard segmentation techniques (e.g., graph cutting [15], dense conditional random field [DenseCRF] [16,17]) or additional model parameters (e.g., boundary prediction [18] and adversarial training [19]). .
The existing scribble-supervised methods have shown the possibility of reducing manual effort in generating training labels, but their adaptation in cell segmentation has not been investigated yet.
Motivation
Contribution
Cell Segmentation
Weakly-supervised Cell Segmentation
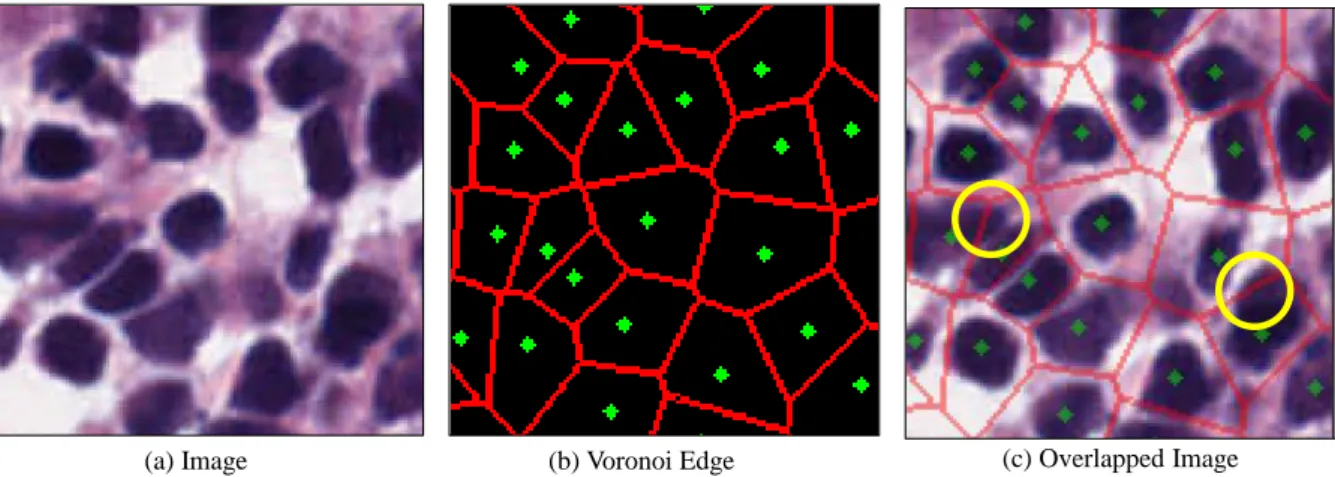
In (c), some Voronoi Edge parts are present in elongated cells. a) Image (b) Full label (c) Dotted label (d) Scribble label Figure 4: Example of different types of labels.
Scribble-supervised Learning
The proposed method is motivated not only to reduce computational time and cost, but also to make good use of the semantic information from scribbles.
Semi-supervised Learning
A conventional pseudolabel can be a biased distribution based on a particular state in the network that differs from the actual ground truth. In this thesis, Scribble2Label proposes a pseudo-labeling method that participates in prediction consistency. The actual scribble thickness used in our experiment was 1 pixel, but in this image it is expanded to 5 pixels for better visualization.
The input sources for our method are the image x and the user-given scribbles s (see Figure 6). Here, the given scribbles are marked pixels (indicated as blue and yellow for the foreground and background, respectively), and the rest of the pixels are unmarked pixels (indicated as black). For unlabeled (unscribbled) pixels, our network automatically generates reliable labels using the exponential moving average of the predictions during training.
The first stage is an initialization (i.e. warm-up stage) by training the model using only the loss of projected pixels (Lsp). After the model is initially trained in the warm-up stage, the forecast is iteratively refined with both planned and unplanned losses (Lsp and Lup). In the following, Lsp is calculated using the scribble annotation, and Lup is calculated using the filtered pseudo-tag.
Warm-Up Stage
In the ensemble stage, an unenlarged image is used for input to the network and EMA is applied to these predictions. Moreover, in the supervised scribble setting, we cannot merge predictions when the best model is found, as in [35], because the given label is not fully labeled. To achieve the valid ensemble and reduce computational costs, the forecasts are averaged every γ epoch, where γ is the ensemble interval.
Learning with a Self-Generated Pseudo-Label
Datasets
Implementation Details
Results
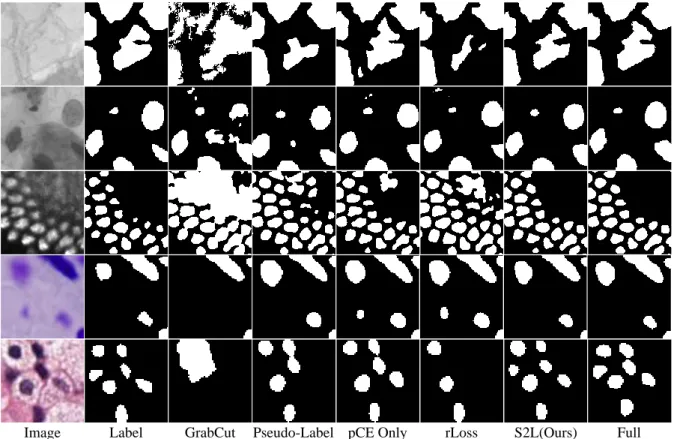
However, due to learning using only scribbles, the method failed to correctly predict the boundary as accurately as our method. rLoss [17] outperformed most previous methods, but our method generally showed better results. The target data were DSB-Fluo, and various amounts of scribbles, ie. and 100% of the skeleton pixels extracted from the full segmentation labels (masks), are automatically generated. What is interesting is that most of the inconsistent pixels are located at the edge of the cell.
Raviv, "Microscopy cell segmentation via convolutional LSTM networks," in 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019). Metaxas, "Multi-scale cell instance segmentation with keypoint graph based bounding boxes," in International Conference on Medical Image Computing and Computer-Assisted Intervention. Biseet al., “Weakly supervised cell instance segmentation by propagation from detection response,” in International Conference on Medical Image Computing and Computer-Assisted Intervention.
Metaxas, “Weakly Supervised Deep Nuclei Segmentation Using Point Labels in Histopathology Images,” in International Conference on Deep Learning Medical Imaging, 2019, p. Paeng, “PseudoEdgeNet: Nuclei Segmentation only with Point Annotations,” in International Conference on Medical Imaging Computing and Computer-Assisted Intervention. Sun, “Scribblesup: Scribble-supervised convolutional networks for semantic segmentation,” in Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2016, p.
Boykov, “On regularized losses for weakly supervised cnn segmentation,” in Proceedings of the European Conference on Computer Vision (ECCV), 2018, p. Zhang, “Guided boundary detection: a scribble-supervised semantic segmentation approach,” in Proceedings of the 28th International Joint Conference on Artificial Intelligence. Sun, “Deep residual learning for image recognition,” in Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2016, p.
Kwak, "Learning pixel-level semantic affinity with image-level supervision for weakly supervised semantic segmentation," i Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2018, s. Byun, "Learning texture invariant repræsentation for domain adaptation of semantic segmentation,” i Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, 2020, pp. Brox, “U-net: Convolutional networks for biomedical image segmentation,” i International Conference on Medical image computing and computer-assisted intervention .
Yvon, »Learning the structure of variable-order crfs: a finite-state per-spective«, v Proceedings of the 2017 Conference on Empirical Methods in Natural Language Processing, 2017, str. Yoo, »Cutmix: Regularization strategy to train strong classifiers with funkcije, ki jih je mogoče lokalizirati,« v Proceedings of the IEEE International Conference on Computer Vision, 2019, str.
![Table 1: Quantitative results of various cell image modalities. The numbers represent accuracy in the format of IoU[mDice].](https://thumb-ap.123doks.com/thumbv2/123dokinfo/10495996.0/28.892.111.800.193.608/table-quantitative-results-various-modalities-numbers-represent-accuracy.webp)
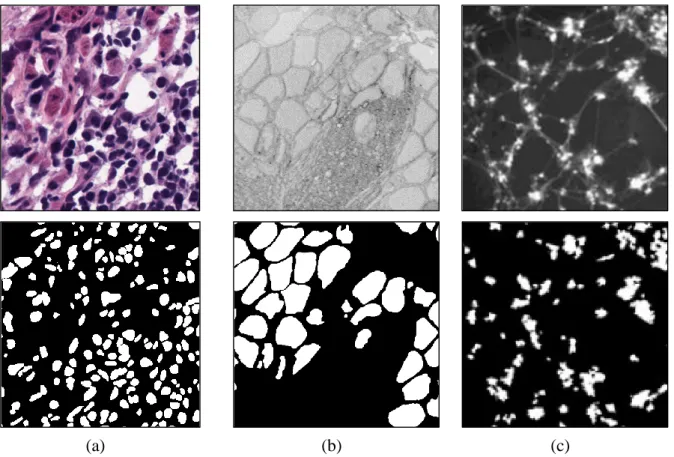
![Figure 2: A sample image and label from [4]. There are 1,309 cells in this single 1,000 x 1,000 pathology image.](https://thumb-ap.123doks.com/thumbv2/123dokinfo/10495996.0/14.892.118.789.713.1055/figure-sample-image-label-cells-single-pathology-image.webp)
![Table 2: Quantitative results using various amounts of scribbles. DSB-Fluo [5] was used for the evaluation](https://thumb-ap.123doks.com/thumbv2/123dokinfo/10495996.0/29.892.167.722.194.551/table-quantitative-results-using-various-amounts-scribbles-evaluation.webp)