LITERATURE REVIEW 5
Medicinal Plants 5
Medicinal plants can cause harmful effects when used improperly, so both the raw material and the finished product must be identified (Suesatpanit et al., 2017). However, there is no scientific evidence to support the effectiveness of medicinal plants used in traditional medicine. Those medicinal plants (whole or part of the plant) with medicinal properties are called natural crude drugs or of biological origin when they exhibit therapeutic properties (Josephine Ozioma and Antoinette Nwamaka Chinwe, 2019).
Although medicinal plants have a long history of effective use, they are still not mainstream medicine.
Identification Methods 7
Traditional Method 8
- Herbarium Voucher 9
In 2017, Begue and researchers stated that the most successful approach for accurate identification of medicinal plants is a manual-based method based on morphological features. Herbarium slips have been used in science since 1556 and are designed as official and permanent records of dried medicinal plants using thick sheets of paper bound together and held vertically (Stearn, 1971). It lays the foundation for plant illustration and provides a permanent record that confirms the occurrence of species at a given locality and time (Deo et al., 2017).
It is known that identification of medicinal plants is often done manually by herbarium taxonomists using a taxonomic guide (Herdiyeni et al., 2013).
Molecular Method 11
- Development of DNA Barcoding 12
- DNA Barcoding of Medicinal Plant 13
- Universal Barcodes for Medicinal
- Ribulose 1,5-biphosphate carboxylase
- Maturase K (matK) 16
- Internal transcribed spacer (ITS) 16
The rbcL gene region is used for DNA barcoding because of its universality, ease of amplification, and comparability ( Hollingsworth et al., 2016 ). However, rbcL is considered modest in discriminating at the species level (Arolla et al., 2015). The matK gene has been widely used as a barcode in angiosperm plants (Yu et al., 2011).
ITS has been developed as an effective universal region for molecular identification of medicinal plants (Samsuddin et al., 2012).
Strategies for Detected Plant Metabolites 17
Metabolite Fingerprinting 18
In addition, it is also used as a screening tool to distinguish samples from different biological sources (Ellis et al., 2007).
Metabolite Profiling 18
Data obtained from the field notebook as shown in Table 4.1 was used to prepare labels for the voucher samples. Data obtained from the field notebook as shown in Table 4.2 was used to prepare labels for the voucher samples. Data obtained from the field notebook as shown in Table 4.3 was used to prepare labels for the voucher samples.
Data obtained from the field notebook as shown in Table 4.4 was used to prepare labels for the voucher samples.
Analytical Techniques- Metabolite
Gas chromatography coupled to mass spectrometry (GC-MS) was realized in the 1950s with commercial instruments in the 1970s (Pitt, 2009). Today, modern LC-MS setups offer a superior choice for structural elucidation of unknown metabolites, namely accurate mass determination and elemental composition analysis (Xie et al., 2008). There is a design that combines LC-MS and metabolite profiling can detect secondary metabolites, including functional compounds in food and medicinal plants (Tamura et al., 2018).
Recently, LC-MS has been shown to be a new powerful technique to identify unknown components in plant extracts through the efficient separation capability of high-performance liquid chromatography and the accurate structural characterization of mass spectrometry (Yang et al., 2009; Kumar, 2017).
Classification of Metabolites 23
- Terpenes 24
- Phenolic compounds 25
- Nitrogen-containing compounds 26
Data obtained from the field notebook as shown in Table 4.5 was used to prepare labels for the voucher samples. Data obtained from the field notebook as shown in Table 4.6 was used to prepare labels for the voucher samples. Data obtained from the field notebook as shown in Table 4.7 was used to prepare labels for the voucher samples.
The data obtained from the field notebook as shown in Table 4.8 was used to prepare the labels for the voucher samples. The data obtained from the field notebook as shown in Table 4.9 was used to prepare the labels for the voucher samples. The data obtained from the field notebook as shown in Table 4.11 was used to prepare the labels for the voucher samples.
The data obtained from the field notebook as shown in Table 4.24 was used to prepare the labels for the voucher specimens. The data obtained from the field notebook as shown in Table 4.28 was used to prepare the labels for the voucher specimens. The data obtained from the field notebook as shown in Table 4.29 was used to prepare the labels for the voucher specimens.
The data obtained from the field notebook as shown in Table 4.30 was used to prepare the labels for the voucher samples. The data obtained from the field notebook as shown in Table 4.31 was used to prepare the labels for the coupon samples. Data obtained from the field notebook as shown in Table 4.32 was used to prepare the labels for the voucher samples.
The data obtained from the field notebook as shown in Table 4.33 was used to prepare the labels for the voucher specimens. The data obtained from the field notebook as shown in Table 4.34 was used to prepare the labels for the voucher specimens. The data obtained from the field notebook as shown in Table 4.35 was used to prepare the labels for the voucher specimens.
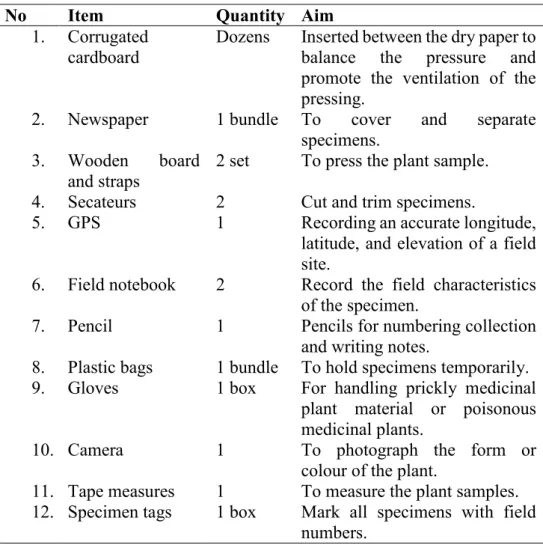
MATERIALS AND METHODS 27
Selection and Collection of Medicinal Plant 27
Prior to the excursion, the herbalist was given a list of 153 local medicinal plants. A field trip with a herbalist was then conducted to collect specimens of each medicinal plant species. The purpose of the field trip was to collect a total of 35 species of medicinal plants from 153 listed species (Table 3.1).
For a quick overview of the experimental design in this study, the experimental layout of this study is shown in Figure 3.1.
Macroscopic Photography 34
The camera was used to photograph the external characteristics of the desired medicinal plants such as leaves, stems, fruit or the whole plant at the time of collection. Plant identification of the 35 selected species was still in the preliminary stages of the plant collection process. Dr Richard Chung, the plant taxonomist responsible for description, identification, nomenclature and classification (Simpson, 2019) from the Malaysian Forest Research Institute was later consulted on the taxonomic identification of medicinal plants.
All data collected were in accordance with the field record suggested by Forest Research Institute Malaysia.
Herbarium Voucher Preparation and Deposition 35
DNA Barcoding 36
- DNA Extraction 36
- Polymerase Chain Reaction (PCR)
- DNA Sequence Analysis 38
Data obtained from the field notebook as shown in Table 4.10 were used to create labels for the voucher samples. Data obtained from the field notebook as shown in Table 4.12 were used to create labels for the voucher samples. Data obtained from the field notebook as shown in Table 4.13 were used to create labels for the voucher samples.
The data obtained from the field notebook as shown in Table 4.14 was used to prepare the labels for the voucher samples.
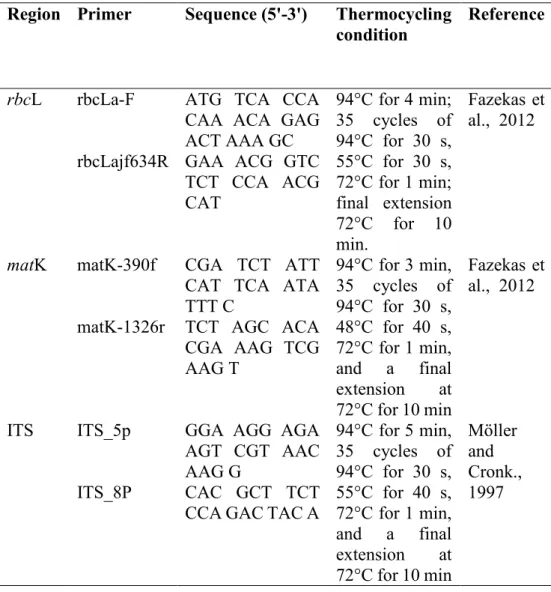
LC-MS/MS 39
- Metabolite Extraction 39
- LC-MS/MS Analysis 40
- Metabolite Identification 41
RESULTS 42
Plant Sampling, Photography and Herbarium
- Andrographis paniculata (Burm.f.) Wall.ex
- Barleria lupulina Lindl. LYMOOI 036 45
- Clinacanthus nutans (Burm.f) Lindau
- Gendarussa vulgaris Nees. LYMOOI 041 51
- Rhinacanthus nasutus (L) Kurz
- Ruellia simplex C. Wright LYMOOI 056 55
- Strobilanthes crispus Blume LYMOOI 033 57
- Catharanthus roseus (L) G. Don
- Alocasia macrorrhizos (L.) G.Don
- Rhaphidophora decursiva (Roxb.) Schott
- Typhonium flagelliforme (Lodd.) Blume
- Calotropis gigantea (L.) W.T.Aiton
- Gynostemma pentaphyllum (Thunb.) Makino
- Cycas revoluta LYMOOI 053 71
- Kyllinga brevifolia Robbt LYMOOI 005 73
- Dioscorea bulbifera (L) LYMOOI 018 75
- Ocimum basilicum L. LYMOOI 040 77
- Orthosiphon aristatus (Blume) Miq
- Vitex trifolia L. LYMOOI 006 81
- Mentha spicata L. LYMOOI 004 83
- Plectranthus amboinicus (Lour.) Spreng
- Punica granatum L. LYMOOI 070 87
- Hibiscus mutabilis L. LYMOOI 045 89
- Urena lobata (L.) LYMOOI 008 91
- Clidemia hirta (L.) D. Don LYMOOI 011 93
- Plantago major L. LYMOOI 034 97
- Morinda citrifolia L. LYMOOI 031 99
- Oldenlandia auricularia LYMOOI 015 101
- Oldenlandia corymbosa (L) LYMOOI 066 103
- Oldenlandia diffusa (Willd.) Roxb
- Lantana camara L. LYMOOI 035 107
- Phyla nodiflora LYMOOI 001 109
- Stachytarpheta jamaicensis (L) Vahl
Data obtained from the field notebook as shown in Table 4.15 was used to prepare labels for the voucher samples. Data obtained from the field notebook as shown in Table 4.16 was used to prepare labels for the voucher samples. Data obtained from the field notebook as shown in Table 4.17 was used to prepare labels for the voucher samples.
The data obtained from the field notebook as shown in Table 4.18 was used to prepare the labels for the voucher specimens. The data obtained from the field notebook as shown in Table 4.19 was used to prepare the labels for the voucher specimens. The data obtained from the field notebook as shown in Table 4.20 was used to prepare the labels for the voucher specimens.
Data obtained from the field notebook as shown in Table 4.21 was used to prepare labels for the voucher samples. Data obtained from the field notebook as shown in Table 4.22 was used to prepare labels for the voucher samples. Data obtained from the field notebook as shown in Table 4.23 was used to prepare labels for the voucher samples.
A mounted voucher herbarium specimen (Fig. 4.24c) with label details in the lower right corner was deposited at the Perdana Botanic Gardens, Kuala Lumpur. The data obtained from the field notebook as shown in Table 4.25 was used to prepare the labels for the coupon samples. The data obtained from the field notebook as shown in Table 4.26 was used to prepare the labels for the voucher samples.
The data obtained from the field notebook as shown in Table 4.27 was used to prepare the labels for the voucher samples.

DNA Barcoding Analysis 113
- Universality of Primer Sequences 113
- DNA Barcoding: Identification Efficiency 113
Untargeted Metabolite Profiling 121
- Andrographis paniculata (Burm.f.)
- Barleria lupulina Lindl. LYMOOI 036 127
- Clinacanthus nutans (Burm.f) Lindau
- Gendarussa ventricosa (Wall.) Nees
- Gendarussa vulgaris Nees. LYMOOI 041 140
- Rhinacanthus nasutus (L) Kurz
- Ruellia simplex C. Wright LYMOOI 056 145
- Strobilanthes crispus Blume LYMOOI 033 150
- Catharanthus roseus (L) G. Don
- Alocasia macrorrhizos (L.) G.Don
- Typhonium flagelliforme (Lodd.) Blume
- Calotropis gigantea (L.) W.T.Aiton
- Gynostemma pentaphyllum (Thunb.) Makino
- Cycas revoluta LYMOOI 053 175
- Kyllinga brevifolia Robbt LYMOOI 005 178
- Dioscorea bulbifera (L) LYMOOI 018 180
- Ocimum basilicum L. LYMOOI 040 187
- Orthosiphon aristatus (Blume) Miq
- Vitex trifolia L. LYMOOI 006 199
- Mentha spicata L. LYMOOI 004 208
- Plectranthus amboinicus (Lour.) Spreng
- Punica granatum L. LYMOOI 070 218
- Hibiscus mutabilis L. LYMOOI 045 221
- Urena lobata (L.) LYMOOI 008 226
- Clidemia hirta (L.) D. Don LYMOOI 011 228
- Melastoma malabathricum L. LYMOOI 009 233
- Plantago major L. LYMOOI 034 236
- Morinda citrifolia L. LYMOOI 031 240
- Oldenlandia auricularia LYMOOI 015 245
- Oldenlandia corymbosa (L) LYMOOI 066 249
- Oldenlandia diffusa (Willd.) Roxb
- Lantana camara L. LYMOOI 035 258
- Phyla nodiflora LYMOOI 001 265
- Stachytarpheta jamaicensis (L) Vahl
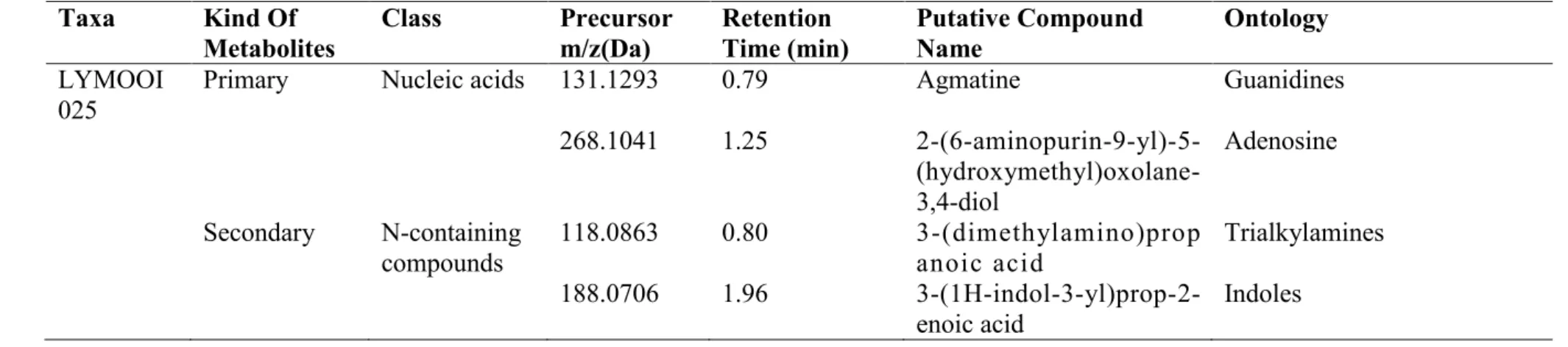
LC-MS/MS analysis of Andrographis paniculata extract allowed the identification of 29 putative compounds (Table 4.39) belonging to different chemical families. LC-MS/MS analysis of Barleria lupulina extract allowed the identification of 26 putative compounds (Table 4.40) belonging to different chemical families. LC-MS/MS analysis of Clinacanthus nutans extract allowed the identification of 17 putative compounds (Table 4.41) belonging to different chemical families.
LC-MS/MS analysis of Gendarussa ventricosa extract allowed the identification of 24 putative compounds (Table 4.42) belonging to different chemical families. LC-MS/MS analysis of Gendarussa vulgaris extract enabled the identification of 9 putative compounds (Table 4.43) belonging to different chemical families. LC-MS/MS analysis of Rhinacanthus nasutus extract allowed the identification of 11 putative compounds (Table 4.44) belonging to different chemical families.
LC-MS/MS analysis of the extract from Ruellia simplex enabled the identification of 28 putative compounds (Table 4.45) belonging to different chemical families. LC-MS/MS analysis of the extract from Strobilanthes crispus enabled the identification of 20 putative compounds (Table 4.46) belonging to different chemical families. LC-MS/MS analysis of the extract from Catharanthus roseus enabled the identification of 35 putative compounds (Table 4.47) belonging to different chemical families.
LC-MS/MS analysis of the extract from Alocasia macrorrhizos enabled the identification of 7 putative compounds (Table 4.48) belonging to different chemical families. LC-MS/MS analysis of Rhaphidophora decursiva extract enabled the identification of 14 putative compounds (Table 4.49) belonging to different chemical families. LC-MS/MS analysis of the extract from Typhonium flagelliforme enabled the identification of 5 putative compounds (Table 4.50) belonging to different chemical families.
LC-MS/MS analysis of extract from Calotropis gigantean allowed the identification of 26 putative compounds (Table 4.51) belonging to different chemical families.
DISCUSSION 276
Identification of 35 Local Medicinal Plants
Identification of 35 Local Medicinal Plants
- Universality of the Three Candidate Barcodes 278
- Discriminatory Rate of Three Candidate
- Factors Influencing the Rate of
Putative Compounds of 35 Local Medicinal Plants 284
CONCLUSION 296








