Dr FC Muchadeyi (Co-supervisor). i) the research reported in this thesis, except where otherwise indicated or acknowledged, is my original work;. ii) this thesis has not been submitted in full or in part for any degree or examination at any other university;. iii) this thesis does not contain other persons' data, pictures, graphs or other information, unless it is specifically acknowledged that it comes from other persons;. iv) this thesis does not contain other persons' writing, unless it is specifically acknowledged that it comes from other researchers. 12 Table 2.3: Genes and the traits in livestock they are responsible for, as discovered by GWAS. 21 Table 3.1: The number of SNPs lost to QC parameters and the number of individuals lost.
INTRODUCTION
- Rationale for the research
- Justification
- Aims
- Objectives
- Outline of dissertation structure
It is important to understand the genetic factors that contribute to the occurrence of sub-vital performance in Swakara sheep. This study intends to use genome-wide SNP genotyping using the ovineSNP50 bead chip to understand the genetics of sub-vital performance in Swakara sheep. The incidence of subvital performance in white Swakara sheep is therefore believed to be a result of inbreeding as a product.
LITERATURE REVIEW
- Introduction
- Coat colour and sub-vital performance in Karakul sheep
- Genes controlling coat colour in sheep
- Molecular markers and genomic tools used to study population genetics
- OvineSNP50 beadchip panel
- Linkage disequilibrium
- Haplotype analysis
- Runs of homozygosity (ROHs)
- Signatures of selection
- Genome-wide association studies (GWAS)
- Conclusion
Molecular markers are polymorphisms at the DNA level (Vignal et al. 2002) that may be associated with certain traits. The availability of high-density SNP arrays has enabled the investigation of homozygosity arrays (Zhuang et al. 2012). Microphthalmia Texel sheep, missense mutation on PITX3 involved in vertebral lens formation (Becker et al. 2010).

PERFORMANCE OF THE OVINESNP50 BEADCHIP IN SWAKARA
- Abstract
- Introduction
- Materials and methods
- Animal Populations
- Purification of DNA
- DNA quantification and quality check
- SNP genotyping workflow
- SNP quality control and analysis
- Results
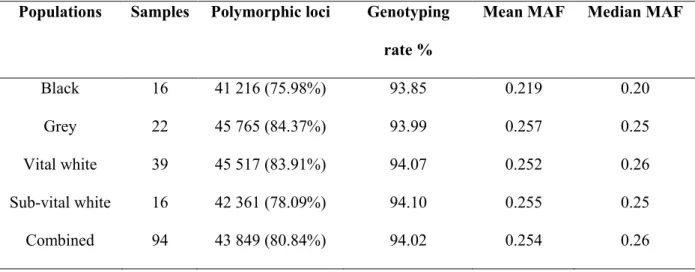
- Genotyping and SNP frequencies pruning
- SNP polymorphisms and MAF
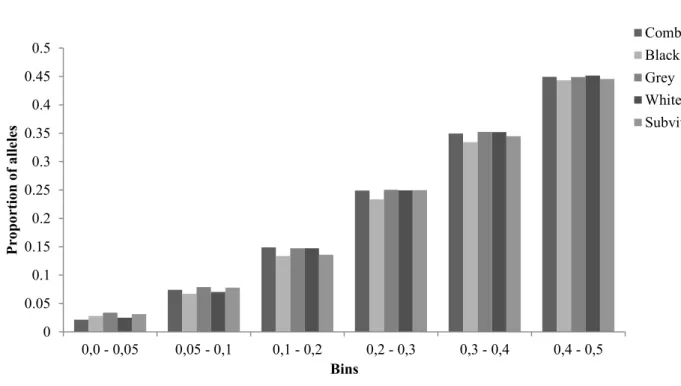
- MAF frequency distribution
- Discussion
- Conclusions
Although the OvineSNP50 has previously been used on many of the world's sheep breeds, it has not yet been used on Swakara sheep. The distribution of SNPs was grouped according to the SNP MAF value in the form of six bin groups. Most markers were lost due to low MAF in the alleles in which the subvital white group lost 11,567 markers, followed by the black subpopulation which lost 10,991 markers (Table 3.1).
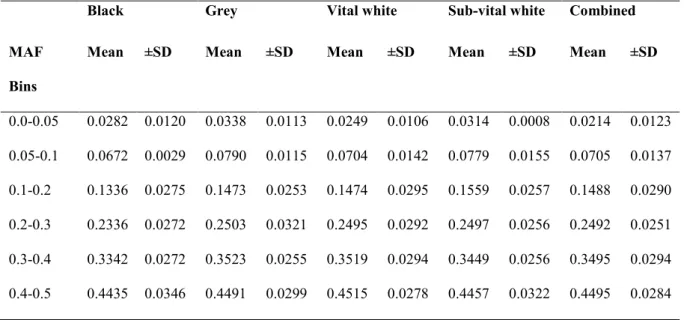
The average MAF in each group was above 0.2 with the highest MAF average of 0.257 in the gray population and the lowest MAF of 0.219 in the black population (Figure 3.2). The bin category that represented informative SNPs was 0.4–0.5, and had a high proportion of SNPs ranging between 0.4435 in the black and 0.4515 in the vital-white subpopulations. Single nucleotide polymorphisms refer to mutations at single base positions in the genome of living organisms (Twyman 2003).
In the current study, the MAF averaged between 0.21 and 0.25, demonstrating the robustness of this genomic tool across a wide range of sheep breeds, including the Swakara. A portion of 80.72% of OvineSNP50 SNPs could be used after imposing QC parameters in the construction of haplotype structures and determination of LD decay in Spanish Churra sheep (Garcia-Gamez et al. 2012). Informative SNPs were also in similar proportions in the subpopulations as represented in the 0.4–0.5 category and had a high percentage of SNPs represented, ranging between 0.4435 (black) and 0.4515 (vital white).
The MAF distribution indicated that the panel is polymorphic and informative in the four color subpopulations of Swakara.

Determining the population genetic substructure of Swakara sheep using
- Abstract
- Introduction
- Materials and methods
- Animal population
- Quality control (QC)
- Expected and observed heterozygosities and inbreeding estimates
- Wright’s fixation indices for determination of population substructure
- Population clustering using Principal component analysis
- Bayesian-based clustering using ADMIXTURE
- Results
- Genetic differentiation
- Fixation indices based on markers per subpopulation
- Population structure within the Swakara population
- Bayesian-based clustering
- Discussion
- Conclusions
A possible accumulation of unique alleles in the white subpopulation may have been the cause of the sub-vital performance observed today. The sub-vital white and gray were also more genetically diverse with an FST value of 0.03215. Genetic differentiation by markers between the white and sub-white groups revealed a SNP on chromosome 10 with an FST = 0.4908 (Figure 4.2).
The population with the highest FST values was the gray versus sub-vital genetic differentiation with an FST value of 0.6846 for a SNP on chromosome 5. The comparison between the sub-vital white and the white subpopulations showed a number of SNPS , but the chromosome with the FST value of 0.4908 was found on chromosome 10. The highest FST per marker was on chromosome 5 (FST = 0.6846) for the gray versus sub-vital white comparison.
The first cluster (A) revealed genetic similarity between the white and subvital groups, consisting of 12 individuals. Higher FST values were observed between gray and white subvital, which had a genetic differentiation FST value of 0.6846 on chromosome 13. These results indicate that the SNP on chromosome 13 is highly differentially selected between subvital white and gray.
The black and the subvital subpopulations shared some genomic regions that differed from the rest of the subpopulations.

Inbreeding levels and distribution of Runs of homozygosity in Swakara
- Abstract
- Introduction
- Materials and methods
- Animals
- SNP genotyping
- Runs of homozygosity
- Identifying potential trait association of SNPs in ROH segments
- Results
- SNP genotyping quality control
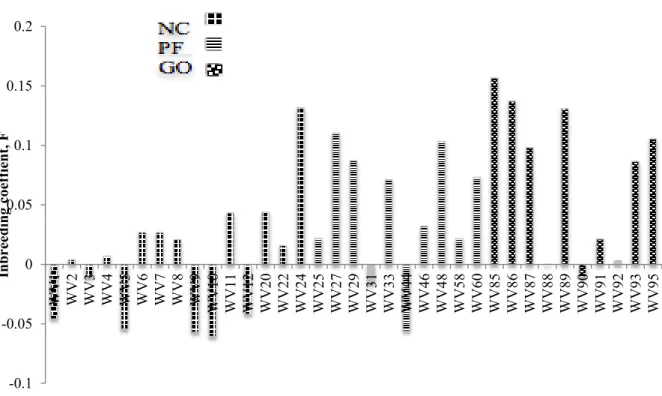
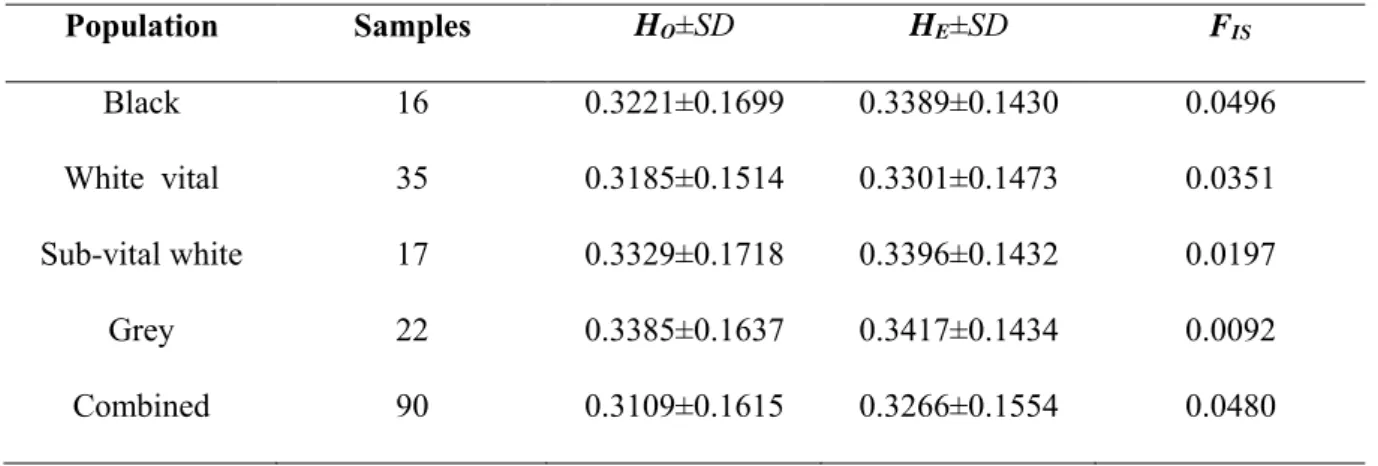
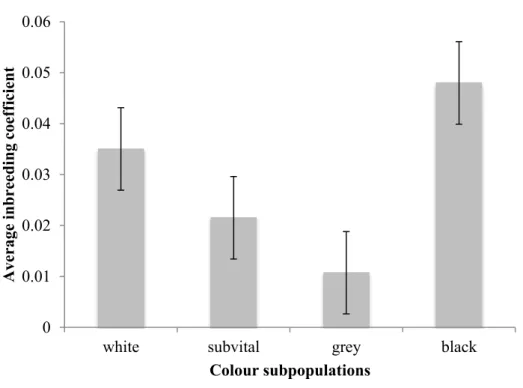
- Inbreeding levels within individuals of the different subpopulations
- ROH prevalence and patterns in Swakara sheep
- Chromosomal coverage of ROH in Swakara population
- Consensus (overlapping) ROHs observed in Swakara population
- Putative function/association of observed ROH
- Discussion
- Conclusions
The production of white Swakara sheep is the product of generations of intensive selection and probably extensive inbreeding, resulting in the accumulation of homozygous recessive alleles. It is hypothesized that the subvital success is the result of inbreeding in the Swakara population, which has led to the accumulation of lethal homozygous alleles in the Caucasian population and not in other skin-colored populations. There is no publicly available information on mating rates in the Swakara lineage.
Understanding the genomic structure and diversity between vital-white, sub-vital white and two other color variants of the Swakara is important in determining the genetics of sub-vital performance to aid Swakara breed improvement programs. The objectives of the study were therefore: i) investigate inbreeding rates in different color subpopulations of Swakara sheep, (ii) examine ROH in the Swakara sheep population using Ovine SNP50K data and. iii) to investigate the associations between ROH and coat color patterns and subvital performance in Swakara sheep. The PLINK group function generated a group of individuals sharing a consensus ROH (cROH) on the same chromosome and within an overlapping region and its distribution in subvital and vital sheep.
Previous studies have linked ROH to economically important traits and this study analyzed ROHs in the context of inbreeding in Swakara subpopulations while determining the patterns of ROH and putative functions. In this study, we sought to find genes or other genetic variants within a screened ROH that may be associated with sub-vital performance in the Swakara sheep using the sheep genome browser and the ENSEMBL genome browsing platform. In addition to having the most ROH segments, BV14 also had the highest FIS value (0.2805) in the black population, showing some correlation between ROH and inbreeding.
The level of inbreeding in the Swakara population was low in subpopulations, with the black population showing the highest level of inbreeding.
GENOME-WIDE ASSOCIATION STUDY OF SUB-VITAL
- Abstract
- Introduction
- Materials and methods
- Animal population, SNP genotyping and quality control
- Genome wide association analyses
- Visualization GWAS data
- Putative functions of SNPs identified
- Results
- The distribution of genotyped SNPs using QQ plots
- Genome-wide association analysis
- Putative biological functions of SNPs identified
- Discussion
- Conclusion
This was based on the hypothesis that sub-vitality only affects some individuals of the white fur color subpopulation and therefore the genetic variant(s) will occur in high frequencies in the sub-vital population. It was therefore worth noting the differences between the normal white individuals and the sub-vital white individuals in a GWAS study. The sub-vital white versus vital white GWA was tested and the results were visualized using Manhattan plot (Figure 6.4).
The production of white fleece from Swakara lambs is hampered by the presence of a sub-vital factor that has a major impact on the white fleece market due to the loss of white lambs and reduced production. These three SNPs also occurred at high frequencies in the subvital white cases compared to the controls (Table 6.2). The high frequencies of the two SNPs (OAR3_s24957.1 and OAR on chromosome 3 in the sub-vital sheep make it more likely that they influence sub-vital performance in white Swakara.
The study also showed that sheep homozygous recessive at the English spotting locus (white) were sub-vital individuals (Fontanesi et al. 2014). The symptoms of sub-vital performance in the English spotting locus in the checkered giant rabbit are similar to those in the Swakara population. The GWAS study identified SNPs associated with sub-vital performance when the analysis was performed using all color subpopulations.
However, GWAS alone was not conclusive as no significant SNPs were identified between the white sub-vital and white vital sheep.
DISCUSSION AND CONCLUSIONS
The genotyping success of the OvineSNP50 beadchip in the Swakara breed demonstrates that the beadchip is highly transferable to the breed and is useful in the study of population genetics in several breeds of interest. Extensive inbreeding and selective breeding reduce genetic diversity, resulting in the accumulation of lethal recessive alleles in populations (Verweij et al. 2014). The frequency of the two SNPs on chromosome 3 that had highly significant frequencies in the subvital individuals presents them as likely candidates for subvitality performance in white Swakara.
Contrary to expectations, there was a failure to observe significant SNPs in the white-only (WO) group. A lethal genetic factor in Garola lambs - a case study. 2010) Microphthalmia in Texel sheep is associated with a missense mutation in the homeodomain 3 (PITX3) gene, a similar pair. South African Journal of Animal Science 23. 1993) Ultrastructure of myenteric ganglia in the rumen, reticulum, omasum and abomasum of grey, white and black Karakul lambs.
American Journal of Human Genetics 81. 2012) Homozygosity and population history in cattle. 2009) Design of a high-density assay in pigs using SNPs identified and characterized by next-generation sequencing technology. 2003) The role of evolutionary history on haplotype block structure in the human genome: implications for disease mapping. Major factors influencing linkage disequilibrium by analyzing different chromosome regions in different populations: demography, chromosome recombination frequency and selection. 2002) Analysis of diversity and population structure in the bred Lipizzaner horse based on pedigree information. 2004).
A novel nonsense mutation in the DMP1 gene, identified by a genome-wide association study, is responsible for hereditary rickets in Corriadale sheep.