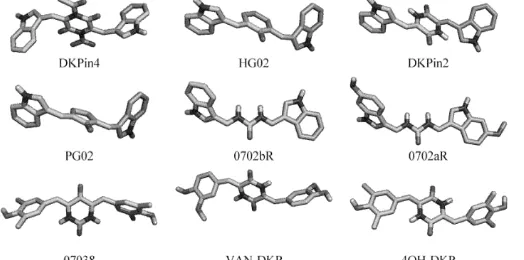
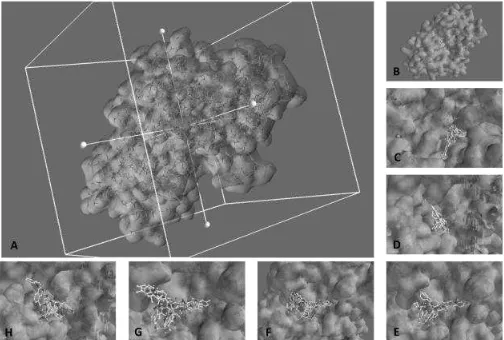
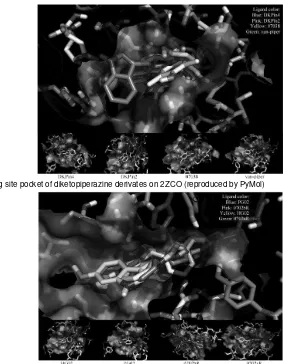
3D-MOLECULAR SCREENING OF DIKETOPIPERAZINE DERIVATES ON Staphylococcus aureus DEHYDROSQUALENE SYNTHASE USING VINA
Teks penuh
Gambar
Dokumen terkait
Keracunan pada manusia disebabkan oleh konsumsi enterotoksin yang dihasilkan oleh beberapa strain Staphylococcus aureus di dalam makanan, biasanya karena makanan
To check binding site between PAF AH (wild type and mutant) and PAF, we performed rigid docking. The affinity energy from this process was -4.0 kcal/mol to
Molecular Docking of Crocus sativus Phytochemicals against Inducible Nitric Oxide Synthase and Phosphodiesterase-9 in Heart Failure Preserved Ejection Fraction.. Heart
Molecular docking results from the active compounds of soursop leaf water extract showed that 5-isopropenyl-3,8-dimethyl-1,2,3,3A, 4,5,6,7-octa hydro azulene (∆G of-6.9 kcal/mol)
Molecular docking analysis showed that the highest binding affinity values for compounds 1 and 2 were to CDK-2 protein with values of −10.14 and −8.8 kcal/mol, respectively.. However,
The results of docking sulphated polysaccharide compounds with the cyclooxygenase-2 enzyme, showed a best binding affinity energy of Gluconic acid ulvan -7.62 kcal/mol similar to the
The docking results using the YASARA structure are binding free energy kcal/mol, the resultant file containing receptor-ligand complex and contact amino acid residues.. The overall
According to the docking results, three compounds are potentially inhibiting LRRK2, namely +-Reticuline with a binding energy of -9.04 kcal/mol and a prediction of inhibition constant