30 Typhoitl Fever and other Salmonellosis Metl
J
InclonesChromosomal rearrangements
in
Salmonella
spp.
Kenneth
E.
Sanderson,
S.L.
Liu*
s2-2
Abstrak
Studi genetik awal memperlihatkan adanya konservasi informasi gen daLam bakteri enterik. Dua metotLe mLLtakhir menggrutakan PFGE untuk menentukan peta fisik genom adalah: ( 1) Digesti parsial dengan endonuklease l-Ceul yang mentototxg DNA bakteri di rrn operon
dari
rRNA (RNA ribosom), urxtuk penetxtuan "kerangka genonikrrn" (ukuran
claLam kb jarak antara operon-operon rRNA). (2) Analisis situs XbaI dan BInl dal.am insersi Tnl0 di kromosom. Urutanfragrnen I-Ceu[ yang adalah ABCDEFG dalam S. typhimuriumlJl2
dan E. coli K-12, ditemukan terkonsenasi pada spesies salmonellayang sebagian besar tumbuhpada berbagai pejamu (pejamtL umum). Akan tetapi pada spesies S. typhi, S. paratyphi C, S. gallinarium dan S. pullorum yang ,nempwtyai pejanu tertentu, fragmen tersebut berbeda sLLswlan karena rekombinasi homolog antara operon rrn menghasilkan translokasi dan inyersi. 127 gaLur S. typhi tlpeIiar mempunyai
2l
urutan berbeda dari fragmen I-CeuI. Inversi d(ux transloknsi ycutg tidak melibatkan operon rnr jarang terdeteksi kecuali untuk inversi pada daerah ZErR (Termination of replicati on). Perubahan penambahan genetik (akibat transfer latercil yang meng-hasilkan insersi DNA tidak homolog) menghasilknn Pathogenicity islands l,ang berisi blok DNA yang memasukkan gen bctru ke gcLlurtertentu, sehingga mendorong terjadinya evolusi cepat dart sifat-sifat baru.
Abstract
Early genetic studies showed conservation of gene order in the enteric bacteria. Two recetxt methods usirtg pul.sed-field gel el.ecto-phoresis (PFGE) to determine the physical map of the genome are: ( I ) partial digestion with the endonuclease l-Ceul,
wlich
digests theDNA of bacteria in the rm operon
for
rRNA (ribosomal RNA), thus estctblishing tlrc "rrn gen.omic skeleton" (the size in kb of the intenalsbetween rRNA operons); (2) analysis of Xbal
antlBlnl
sites withinTnI) insertions
in the cfuontosome. The orderof
I-CeuI fragments, which is ABCDEFG in S. typhimurium LT2 andE. coli K-12, was found to be consen,ed in most Salmonella species, nost of wh.ich grow inmanyhosts(host-generalists).Howeve4 lnS,typhi,S.paratyphiC,S.gallinarum,an.dS. pullorum, specieswhichnrehost-specializecl,these fragments are rearranged, due to homologous recotnbinqtion between the nn operons resulting in transl,ocations an.d inversions. Inversions and translocations not involving the nn operons are seldom detected except
for
im,ersions over th.e TER regiorz (termination of replication). Additive genetic changes (due to lateral transfer resulting in insertion of non-homologous DNA) have resultedh
" patho-genicity islands" containing blocks of DNA which provide new genes to specific strains, thus driving rapid evolution of new traits.Introduction
Gene
orders
in
the
chromosomes
of
enteric
bacteria
such as Escherichia
coli
K-12
and Salmonella
ty-phimurium
IJl2
are
reported
to
be
strongly
con-servedl'2;
the
chromosomes
of
other
genera such
asBacillus
andStreptomyces
arerelatively
unstable3'4.This
extreme
conservation
in
enteric
bacteria
is
sur-prising,
because
during growth
in
culture, and
pre-sumably
in
nature, chromosomes frequently
rear-range; duplications
of
segments
of
the
chromosorne
occur
at
high
frequencies
(10-3to
10-5)s-7,and some
inversions and translocations, especially those with
endpoints
inthe rrn
operons, are commons,8.
The
ob-SaLmonelLa Genetic Stock Centre, Department of Biological Sciences, University of Calgary, Calgary, Alberta, Canada T2N
1N4
*
Departrnent of Medical Biochernistry, University of Calgary, Calgary, Alberta, CanadaT2NlN4
served
stability of
the chromosome
during evolution,
in
spite
of
the high frequency
of
rearrangements in
culture,
indicates
that
major forces
mllst
selectagainst
cells with
rearrangements,
resulting
in
elimi-nation of cells
in
which
the genome has rearranged.
The
purposes
of
this
review
are
to
evaluate
our
pre-sent
knowledge about the
degree
of
stability
of
the genomeof
enteric bacteria,
and to discussthe
mecha-nisms which
have
contributed
to
bacterial evolution
leading
to
virulence, especially
in the
genusSalmo-nella.
Methods
of
Genome
Analysis
Mapping by
genetic methods
such as conjugation andtransduction, used
to
construct
linkage maps
of
E
coliK-12
and S.typhimuriumlJl2,is
relatively
slow,
but
physical methods using
endonuclease
digestion
and
PFGE
permit
rapid
construction
of
genomic
l-Ceul
analysis
The
endonuclease
I-CeuI together
with
PFGE is
es-pecially
suitable
for construction of
agenomic
cleav-age map
showing
thenumber
andlocations
of
the
rrn
genes
for
rRNA (the
"rrn
skeleton")10-12.I-CeuI
isencoded
by
aclass
I
mobile
intron which is
inserted
into the
rrl
genecoding
for
the large subunit rRNA
(23S-rRNA)
in
the chloroplast
DNA
of
Chlamydo-monas
eugamatost3.I-CeuI is specific for
and cutsin
a
19bp
sequenceinThe
rrl
genel4. BecauserDNA
se-quences arestrongly
conserved,
thel-CeuI
site ispre-sent
in
all
seven
rrl
genes
of
enteric
bacteria,
but
atno
other site
so
far
detectedtO.The order
of
I-CeuI
fragments
and
their
approximate sizes
for
S.
ty-phimurium
IJ|Z
are
in
Figure 1A;
the order
is
the samein E.
coli
K-I2
andthe
sizes4e
sipil4ls'16.
{n
addition, fragments that
are adjacent on the
chromo-some can be
determined by
partiall-Ceul digestionll.
Preparation
of high-molecular weight
genomic
DNA,
endonuclease
cleaving
of
DNA
in
agarose blocks,
separation
of
the
DNA
fragments
by
PFGE and
dou-ble cleavage techniques
aredone
asreportedlT'l8.
Di-gestion
by
I-Ceul,
including partial digestion,
usesthe methods described earlierl
l. For partial
digestion,
the enzyme concentration must be diluted;
it
is
im-portant to titer
the enzyme
prior to
use.Tn10 insertions
The
transposon
Tn10 has one site
for
the
endonu-clease
Xbal,
andtwo
sites
for
BlnI
(=AvrII);
thus
in-sertions
of
TnI0
into specific
genes enablesthe
map-ping
of
these genes
on the
physical
chrornosome.Many
genes
which contained Tn10 insertions
were
mapped
on the
chromosome
of
S.
typhimurium
U121'7'19'these transposons
were
transduced
by
bac-teriophage
P22
inTorelated
species such as S.enteri-tidiszÙ,
S.paratyphi
A2t,
S.paratyphi Bzz,
andS
ty-phizz, thus
permitting
the
mapping of
the
location
of
specific
genesby physical
analysis
of
the
location
of
the
sites
for
XbaI
andBlnl.
CHANGES
IN
THE
GENOME
Changes
may be
of
three
different
types:
point
muta-tions
(basepair
changes); genetic rearrangements(dele-tions, duplica(dele-tions, transloca(dele-tions,
andinversions);
ad-ditive
genetic
changes(lateral
transfer due
to
geneticexchange
resulting
in
insertion
of
non-homologous
DNA),
resulting
in "pathogenicity
islands".
Base
pair
changes
These are detectable
by PFGE
methods
only
if
they
occur
in
the
target site
of
the
digesting
enzyme,
where they
result in restriction fragment
length
poly-morphisms (RFLPs) which
can
be
detected
by
most
endonucleases, and
can
give
anapproximate
measureof
the number
of
basepair
changes.However, the
19bp
site
of
I-CeuI
within
the
23S
rRNA
gene
wasnever lost due
to
mutation, and was not formed by
chance
at any site outside the
rRNA
genes (though
other
base
pair
changes
occur, and are
detected
by
nucleotide
sequencing,
or by
other
endonucleases).Genetic rearrangements
In
most
generaand
species, these arerarely
detected
in wild
type strains
of
theenteric bacteria;
this
is
bestillustrated
by
the
fact
that
the
chromosomes
of
S.
4r-phimuriumIJl2
andE.
coli K-12
have retained
simi-larity of
genes and geneorderl'2.
However, two
types
of
genetic
changes aredetected, at least
in
sorire spe-cles.rrn-operon
exchanges
Based on genomic maps
for
the enzymes
Xbal, BlnI,
SpeI,
and
I-CeuI,
rearrangements
due
to.inversions
and
translocations
resulting from
homologous
recom-bination between the
rcn
operons are
very
rare in
most
species;
Thel-Ceul
fragments are
in
the
order
ABCDEFG
in
each
of
the
species
S. typhimuriumls
(Figure
1),
S.paratyphi
822, and S.
enteritidis2], the
same order as
in
E
coli K-l2ts.
Because I-CeuI
cleaves only
rrn
operons and because
the
rrn
skele-ton
is
highly
conserved
in
enteric bacteria,
related
wild
type
strains
usually yield identical fingerprints;
for
example,
seventeenindependent
wild
type
S.
ry-phimurium strains
gave
very similar
I-Ceul
digests,indicating conservation
of
the order
of
\-Ceul
frag-ments on the
chomosomell.
However,
in
someof
the
speciesof Salmonella,
espe-cially
those
which
are
host-specialized, the order
of
I-CeuI
fragments is rearranged, although the order
of
genes
on individual fragments
is
similar23,2a. Theserearrangements
result
from
homologous
recombina-tion
between
pairs
of the
seven
rrn
operons, which
are
repetitive
DNA in
a
chromosome
in
which
most
of
the
DNA
is unique. For example, the order
of
I-CeuI
fragments
in
S.typhi Ty2 (limited
to
growth in
humans)
is
changed
to
AGCEFDB (Figure 2). The
rnr
skeletons
of
127
wild
type
strains
of
S. typhi
showed
21different
orders,which
we
called
"genome
types", postulated to
be dueto
inversions
andtranslo-cations
with rrn
endpoints2s(Figure
3). The
genomesof
strains
of
S.paratyphi A2I
and C26(both
also
Typhoid Fever and other Salmonellosis
(limited
to
growth
in
fowl)
(Liu
and Sanderson,
un-published data).
The
order
of I-CeuI
fragments
in
the
chromosomes
of
other
speciesof
Salmonella, most of
which
are
host-generalists
able
to
grow
in
many
hosts,
is strongly
conserved
(Liu
and
Sanderson,un-published data). Postulated patterns
of
homologous
recombination between
the seven
rrn
operons
areshown
in Figure
4.Other enteric bacteria
also show
some
rearrange-ments.
The
order
of
genes
on
the chromosome of
Klebsiella oxytoca
M5la
resembles S.
typhimurium,
but the chromosome
differs
as
follows:
there
areeight rrn operons instead
of
seven; one
of
thel-Ceul
fragments
is
translocated
to
anew location;
the
total
size
of
the
chromosome
is
5200
kb
rather than 4800
kb as
in
S. typhimurium (Figare
1)zz.tor,nt
orn"t
wild
type
strains
of Klebsiella
spp.
all have
eight
rrn
operons, and in
afew
casesthere
arefurther
translo-cations
of
the I-CeuI fragments
(Liu
and
Sanderson,unpublished
data).Inversions
in
TER region
The
second category
of
genetic reaffangement
in-volves inversions over TER, the terminus region of
the
chromosome.
This
is the
only region in which
in-versions
in wild
type
strains of
Salmonella
are
com-mon (other than inversions involving
rrn
genes, dis-cussedabove).
Inversions overlap
the
TER region in
the
following species, when compared
with
S.
4r-phimurium
(the size
of
the
inversion in kb is
shown):
E.
coli,480
kbls,16. S.enteritidis,
750
p6zo;S.typhi
,500
kb;
S.paratyph, C, 700
kb26.No inversion
wasdetected
in
S.paratyphiB22.
All
theseinversions
dif-fer by
atleast
oneend-point.
The
geneorder
in
S.ry-phimurium
may
beconsidered the
ancestral order, be-causeall
inverted
orders
could be
obtained
in
single
events
from
the S.
typhimurium
order,whereas more
complex
events
would
berequired
from
other
orders,but this may simply reflect high frequency
of
inver-sions
in this region. Inversions
at many
points
in
thegenome are detected
in
experimental studies
in
E
coliK-I2
and S.typhimurium LT2,
though
someend-points are much less common than
efhslsT'28.f\r/6
types
of recombination occur
athigh
frequency
in E
coli
K-12
in
the TER region:
recA-dependent
ho-mologous recombination,
resulting
in
deletions29;site-specific recombination
at
the dif-site in
the
TER
region, due to
activity
of
two
related
recombinasesXerC
and
XerD
of
the lambda
integrase family30.
11is
likely
that
thesetypes
of recombination
arerespon-sible
for
inversion
in wild
type
strains
in
Salmonella
species,though this is not yet
proven.
Med
J
IndonesAddition of new
genes
through
lateral
transfer
This involves entry of non-homologous
genes,
pre-sumably by classical genetic transfer methods
such asconjugation
andtransduction,
andintegration ofthese
genes
into
the chromosome.
This
results
in
segmentsor "loops" of DNA in
onestrain for
which
there is no
homologue
in
closely related
species
or
genera.Loops
of
DNA
distinguishing
E.
coli
and
S.
ty-phimurium were noted by
Riley
and
colleaguss2,3l.
These
loops have
been
called pathogenicity
islands
(though
not
all
such loops may
function'in
patho-genicity);
in
many
cases
they include
a
block of
genescontributing
to
aspecific virulence
phenotype.
Two Salmonella pathogenicity
islands
( SPf
,both of
about
40 kb,
havebeen
identified;
SPI-I
governs
theability of
Salmonella
to
invade epithelial
cells32 andSPI-2
mediates survival
within
macrophages33'34.These
pathogenicity
islands
distinguish different
gen-era
of
enteric bacteria,
but
chromosomes
of different
species
of
Salmonella also
differ
by
loops
of
this
type,
andPFGE analysis
hasidentified
theseloops
by
two
methods.
Firstly,
genetic maps
of
Salmonella
show that intervals between
the same genepair in
dif-ferent
speciesmay be
very different. For
example,
in
S.
typhi the mel-poxA interval is
120
kb longer
than
in S. typhimurium and other
species;
this loop
wasfound to
carry the
viaB
genefor
the Vi
antigen,
miss-ing from most other
Salmonella23,2s.This
suggestedthe existence
of
apathogenicity island; this
was
con-firmed,
for
asite-specific
recombinase3s andthe
syn-thesis
of
type
IV pili36 were shown
to
be controlled
by
genesin
this major pathogenicity island
of S.
ry-phl. Secondly,
changes
in
the length
of
the
l-Ceu-Lfragments indicates insertions or
deletions;
e.g.the
I-CeuI-D fragment is
136kb in
S.typhibut
only
92
kb
in all
other Salmonella
speciesexamined
sofar,
indi-cating
aninsertion
of about
44 kb in this
region23.Pathogenicity
islands have also been
identified in
E.coli strains
of the
enteropathogenic
E
coli
(EPEC)
group
and
the uropathogenic E.
coli
group
(UPEC),
as
well as
in
Yersinia
pestis
and Vibrio
cholerae31.Lateral transfer resulting
in
"loops"
of
species-spe-cific
DNA is
amajor
genetic mechanism
resulting in
species adaptation
in the enteric bacteria, including
SalmonelLa.
These
islands
of
unique
DNA
appear
to
be
responsible
for
the
major differences
in
patho-genicity
between the
closely
related
speciesand
Acknowledgements
We
thank
Lili
Lei
for technical
assistance.The work
was
supported
by
grants
from
the Natural Sciences
and Engineering
Research
Council
of
Canada,
andgrant RO1A134829
from the National Institute of
A1-lergy and Infectious
Diseases
of
the National
Insti-tutes
of Health of
the
USA.
References
1.
Sanderson KE. Genetic relatedness in the family Enterobac-teriaceae. Ann Rev Microbiol 19'16;30: 32'7-49.2.
Krawiec S, RileyM.
Organizationof
the bacterial genome.Microbiol Rev 1990; 54: 502-39.
3.
FonsteinM,
HaselkornR.
Physical mappingof
bacterialgenomes. J Bacteriol 1995; 177 : 3361-9.
4.
Kolsto AB. Dynamic bacterial genome organization. MolecMicrobiol 1997 ; 24: 241-8.
5
Anderson RP, Roth JR. Gene duplication in bacteria:altera-tion of gene dosage by sister chromosome e exchange. Cold Spring Harbor Symp Quantit
Biol
1978; 43: 1083-7.6.
Hill
CW
Harnish BW. Inversions between ribosomal RNAgenes of Escherichia coli. Proc Natl Acad Sci, USA 1981; 78:7069-72.
'7.
Roth JR, Benson N, Galitski T, Haack K, Lawrence JG, Mi-esel L. Rearrangements of the bacterial chromosome: forma-tion and applications. In: eds. Neidhardt FC, Curtiss R III, In-graham JL, Lin ECC, Low KB, Magasanik B, Reznikoff W S, RileyM,
SchaechterM,
Umbarger HE. Escherichia coliand Salmonella typhimurium: cellular and molecular biology American Society for Microbiology Press Washington, D.C. 1996:2256-76.
8.
Hill
CW, Gray JA. Effects of chromosomal inversion on cellfitness in Escherichia coliK-72. Genetics 1988; 119: 771-8.
9.
Smith CL, Econome J, Schutt A, Klco S, Cantor CR. A physi-cal map of the Escherichia coli K-12 genome. Science 1987;236: 1446-53.
10. Liu SL, Hessel A, Sanderson KE. Genomic mapping with
I-Ceul, an intron-encoded endonuclease specific for genes for ribosomal
RNA
in
Salmonella spp, Escherichiacoli,
and other bacteria. Proc Natl Acad Sci, USA 1993;90: 6874-8.11.
Liu
SL, SandersonKF,
l-Ceul reveals conservationof
the genome of independent strains of Salmonella typhimurium.I
Bacteriol 1995; 177: 3355-7 .12. Honeycutt R, Mc Clelland
M,
Welsh J.A
physical map of Rhizobium meliloti 1021. J Bacteriol1993;
175:6945-52. 13. Marshall P, Lemieux C. Cleavage pattern of the homingen-donuclease encoded
by
the fifth intron in the chloroplast subunit rRN{.-encoding geneof
Chlamydomonaseugama-tos. Gene 1997;704
l24l-5.
14. Marshall P, Davis TB, Lemieux
C.'lhel-Ceul
endonuclease:purification and potential role in the evolution of Chlamydo-monas group
I
introns. Eur J Bacteriol 1994;220: 855-9. 15. Bachmann BJ. Linkage map of Escherichia coli K-12,edi-tion 8. Microbiol Revs l99O; 54: 130-97.
16. Berlyn MB, Low KB, Rudd KE. Integrated linkage map of
Escherichia coliK-12, edition 9. In: eds. Neidhardt FC, Cur-tiss R
III,
Ingraham JL,Lin
ECC, LowKB,
Magasanik B, Reznikoff W, Riley M, SchaechterM,
Umbarger HE.Es-cherichia
coli
and Salmonella: cellular and molecular biol-ogy. American Societyfor Microbiology
Washington DC 1996:1715-902.17.
Liu
SL, SandersonK
E. A physical mapof
The Salmonella typhimuriumLI2
genome made by using XbaI
analysis. JBacteriol 1992; 174: 1662-'12.
18.
Liu SL,
HesselA,
Sanderson KE. The Xba l-BIn I-CeuI
genomic cleavage map of Salmonella typhimuriumYl2
de-termined by double digestion, end-labelling and pulsed-field gel electrophoresis. J Bacteriol I 993; l'75: 4104-20.19. Wong KK, Mc Clelland M. A BInI restriction map of the
Sal-monella typhimurium LT2 genome. J Bacteriol 1992; l'14:
1 656-6 1.
20. Lil'r SL, Hessel
A,
Sanderson KE. The Xba I-BIn I-CeuI
genomic cleavage map of Salmonel,la enteritidis shows an in-version relative to Salmonella typhimurium LT2. MolMicro-biol 1993; 10: 655-64.
21. Liu SL, Sanderson KE. The chromosome of SaLmonella pa-ratyphi
A is
invertedby
recombination betweenrrnH
andrrnG. J Bacteriol 1995; 177: 6585-92.
22. Liu SL, Hessel A, Cheng HYM, Sanderson KE. The Xba
l-BIn I-Ceu
I
genomic cleavage map of Salmonella paratyphi B. J Bacteriol 1994; l'76(4): 1014-24.23. Lht SL, Sanderson KE. The genomic cleavage map of Sal-monelLa typhi Ty2. J Bacteriol 1995
;
17'7 : 5099- I 07. 24. Liu, SL, Sanderson KE. Rearrangements in the genome of thebacterium SaLmonella typhi. Proc Natl Acad Sci USA 1995;
92: 1018-22.
25.
Liu
SL, Sanderson KE. Highly plastic chromosomal organi-zationin
Salmonella typhi. Proc Natl Acad Sci USA 1996;93: 10303-8.
26. Hessel A, Liu SL, Sanderson KE. The chromosome of
Sal-monella paratyphi C contains an inversion and is rearranged
relative to S. typhimurium l-i12. Abstr Ann Mtng. Amer Soc
Microbiol 95û Annual Meeting 1995; 503.
27. Llu SL, Qi CPF, Stewart
V
Sanderson KE. A genome map ofKlebsiella oxytoca M51a. Ann Mtng. Am Soc Microbiol 97d' Annual Meeting 199'l ; 319.
28. Mahan MJ, Segall AM, Roth JR. Recombination events that reârrange the chromosome: barriers
to
inversion.In:
eds. Drlica K, Riley M. The bacterial chromosome (AmericanSo-ciety for Microbiology Washington DC 1990; 341-9.
29. Louarn J, Cornet
fl
Francois V, Patte J, Louarn JM. Hyperre-combinationin
the terminus regionof
The Escherichia colichromosome: possible relation
to
nucleoid organization. JBacteriol 1994; 17 6: 7 524-31.
30. Cornet
fl
Louarn J, Patte J, Louarn JM. Restriction of the ac-tivity of the recombination sited,/to
a small zone of theEs-cherichia coll chromosome. Genes Dev 1996;
l0:
1152-61 .Es-34
32
Typhoid Fever and other Salmonellosis
cherichia
coli
and Salmonella typhimurium.In:
eds. DrlicaK,
Riley M., The bacterial chromosome. American Society for Microbiology Washington DC 1990; 85-95.Mills
DM,
BalajY
Lee CA.A
40 kilobase chromosomal fragment encoding Salmonella typhimurium invasion genesis absent from the corresponding region
of
the Escherichia coli chromosome. Mol Microbiol 1995;l5:
749- 59. Ochman H, Soncini FC, Solomon F, Groisman EA. Identifi-cation of a pathogenicity island required for Salmonella sur-vival in bost cells. Proc Natl Acad Sci USA 1996;93:7800-4.Shea JE, Hensel M, Gleeson C, Holden DW. Identification of
a virulence locus encoding a second type
III
secretion systemin Salmonella typhimurium. Proc Natl Acad Sci USA 1996; 93: 2593-7.
ZhangXL, Morris C, Hackett J. Molecular cloning, nucleo-tide sequence, and function of a site-specific recombinase
en-Med
J
Indonescoded in the major "pathogenicity island" of Salmonella ty-phi. Gene in press 1997
36. Zhang
XL.
Characterization of a new site-specific recombi-nase and a new type IVJike pilin operon encoded in thema-jor
"pathogenicity island"of
Salmonella rypfti. PhD thesisHong Kong University of Science and Technology 1997 37. Groisman
EA,
OchmanH.
Pathogenicity islands: bacterialevolution in quantum leaps. Cell 1996;87:791-4.
38. Noller HF, NomuraM. Ribosomes. In: eds. Neidhardt FC, In-graham JL, Low KB, Magasanik B, Schaechter M, Umbarger HF,. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. American Society for Microbiology Washington DC 1987; 104-25.
39. Thong KL, Puthachearg SD, Pang T. Genome size variation among recent human isolates of SaLmonella typhi. Res
Mi-crobiol 1997; 148: 229-35.
B
JJ
34
35
A
'a^'a-Ôn
,,1*
è
s
rrnD
rtn D
[image:5.595.43.542.75.674.2]rrnDf E
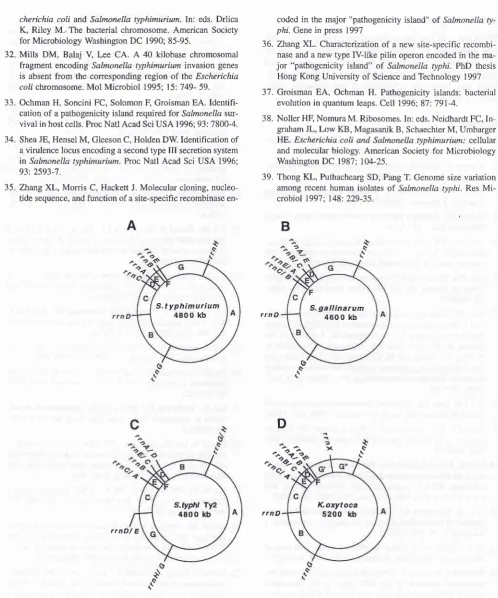
Figure 7. The
"nn
genomic skeleton" of the chromosomes of S. typhimurium LT2 (A), as reported earlierrs, of S. gallinarum (B) (un-published data), of S. typhi Ty2 (CSzt-zs, and of Klebsiella oryîoca (D) (Liu SL and Sanderson KE, unpublished data) The letters A to G indicate the maporde
obtained by digestion with thee
The rrn operon designation at the site of l-Ceuldigestion is based
on
rtc data reported earliertsfor
S.
ther species hybridrm
operons are named ac-cording to postulated homologous recombination between rrn operons, or are named according to location. Each ofthe species has seyenrrn operon except
for
K. orytoca, which also has an eighth rrn operon designated rrnX.o
f
D
+
s t\
5
x
S.typhimurium
4800 kb
S.gallinarum
4600 kbK.oxytoca
5200
kb S.typhlty2
4800 kb
à^;
Ë:t
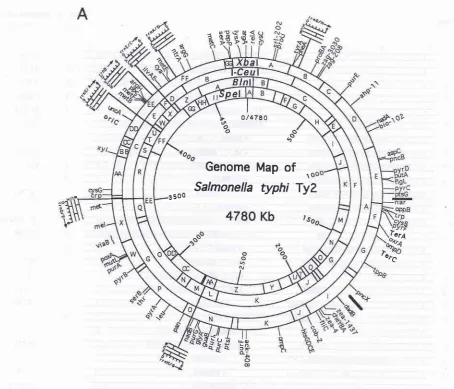
Figure 2 A, Genomic cleavage map of S. typhi Ty2
for
XbaI, I-Cezz
rrs(l6S
5097 154 1982
788
o,
Genome Map
of
1
-
Salmonella
typhi
TyZ
3 500
[image:6.595.55.533.594.696.2]36
Typhoid Fever and other SalmonellosisType
l*
2
3
4
5
6
7
B
9**
10
.l
1
12
13
14 15 16
17
'1
B
I9
?o
71
72
)2
z4
z5
?6
E. coli
KlZ
S. en
SpB
S. tm
Lr2
Med
J
IndonesNo.
of
Representativestrains
strain2
22
57
5
2
B
2
4
0
2
0
7
2
0
1 1
z
4
0
0
?
2
2
1 1
26T3 26Tl ?6T4 26T8
2611 9 26112 CC6
26T50
Tv2
26r.49
cDc3B"-82
26T3 B
26T9
ln4
25r35
?6T40
417ry
26156 26T32
SARB63
P127566
nn'A
-+-> -+
rrnH-->
rrnE
[image:7.595.54.541.96.576.2]-+
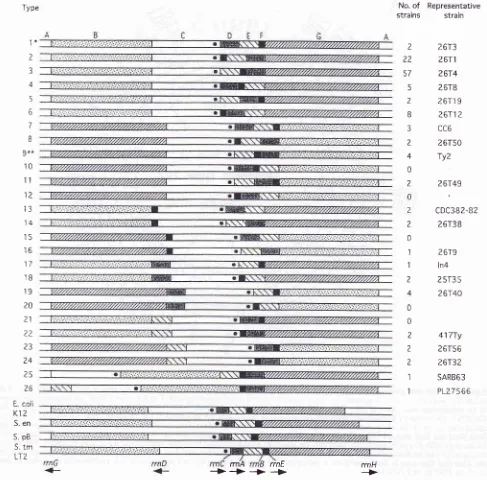
Figure
3'
The order of I-CeuI fragments in 126 stains of S. typhi. The order infour
species which show very little variability withineach species is also shown, E. coli K-12, S. enteritidis (E.en), S. paratyphi B (S,pb), and S. typhimurium LT2 13.tmffZ1. The siies in kb
of the fragments in most strains of S. typhi are at the top; the lengths of the intervals are approximately to scale, The order of I-CeuI
fragments B to G was determined by partial digestion by the endonuclease l-Ceul, and sepàiation of the Tragments by putsed.-field get
electrophoresis. The I-CeuI-A fragment (2400 kb in size) is inferred to
join
the left entl to the right"ia
oyilrn-Jr"gmenis'shown,'forminga circle, but its orientation is not known. The orientation of l-Ceul fragments B, D, E,
4
and G can be inferied from the polarity of themt
genes, but I-CeuI-C could be inverted. The solid dot in the I-CeuI-C fragment is the proposecl locatiàn of oriC; this is known-in E.coli K-12 and S. typhimurium LT2, but is inferred in the other strains. The number of strains'of each genome-type is shown; some of the
A.
Duplication
A*
B
*G.-*D*E*Fr G.-A
"'"'."'.rj.-,ir..
.r:i1...
a
I .-|->-ùt
-fr
A- B * c.
-F
-D-E--
G
*A
A. G +C. lDrEfFD B
pA
+
+
+
D.
lnversion
A*
B
*C.J-E-F-- G-rA ---ù
tt
rrnD
rrnE