A
putative localization
of the
ureaplnsma
urealyticum
IgAl
protease
Felix
Melchizedech Mesak, Nana
SuhanaAbstrak
Salah satufaktor patogenisitas (Jreaplasma urealyticumialah protease
IgAl.
Visualisasi aktivitas protease IgAI dilakukan dengan leknik SDS-PAGE yang memperlihatlan dua fragmen hasil pemecahan rantai beratIgAI manusia yaitu Fc dan Fab masing-masing
berukuran 35 dan 32kDa.
Aktivitas protease hanya terdapat pada sel ureaplasma. Kami tidak dàpat menentukan adanya aktivitas dalam supernatan hasil senftifugasi kultur cair ureaplasma. Produk aktivitas IgAI baru dapat di'detelcsi setelah tiga jam inkubasiberdasarkan cara uji yang kami lakukan. Sedangkan ekstraksi dan partisi fase sel ureaplasma dengan Trtton X-100 dan Triton X-l 14 memperlihatkan kemungkinan pendewasaan protease
IgAl di dalam sitosol sebagai
protein terlarut dan atau pada akhirnya dieksposisi sebagai enlim teikat membran. Dengan memanfaatkan piranti lunak Corel Photo Paint 7, densitas derajat abu-abu dan jumlah'piksel pita-pita berbeda hasil SDS-PAGE dapat dihitung dan menghasilkan nilai tertentu yang disebut skor intensitas pita. Skor ini digunakan sebagai pembanding terhadap parameter uji aktivitas protease IgAl di berbagaifraksi sel ureaplasma dan visualisasinya dalam bentuk grafik linier atau diagram sebar.Abstract
One of the Ureaplasma urealyticum pathogenicity factors is IgAl protease. IgAl protease activity was visualized
by SDS-pAGE
showing two fragments of digested heavy chain ofIgAt with 35 and 32 kDa in sizes denoted as Fc and
Fab, respectively.'The proteaseactivity was only found
in ureaplasma
cells.
We could not determineany activity
in the supernatant
after removingthe cells by
cenÛifugarton. Under our test condition, the products of the IgAl
activity were d.etected after three hours. Based on Triton X-100 extraction and Triton X-I14 phase partitioning,IgAl protease is synthesized to mature
in the cytosol as a soluble protein and./or to befinally exposed as a membrane bound-enzyme. Employing the advantage of Corel Photo Paint 7 sofnvare, the density of grayscales and pixels of distinct bands of the SDS-PAGE were evaluated and these values used to establish a band intensity score. The Score is used as a comparison tool toward testing parameters of the enzyme activity in cellular
fractions,
They are visualized in linear graphs orscatter diagrams.
Keywords :
IgAl, IgAl
protease, ureaplasma, pixels, TritonX-100, TritonX- j14.Ureaplasma urealyticum
sized about
330 nm
in
diameter,
is
one
of
the smallest self-replicating
or-ganisms.
This
bacterium
lacks
a
cell
wall.
It
is
microaerophilic with
a growthoptimum
at37oC
and apH
of
6.0.'
This microbe mostly colonizes
humanurogenital tracts
of adults.
Its
presencein
these tractsmay lead
to
diseases
such as: urethritis and
stoneformation, prostatitis,
infertility,
funisitis,
endo-metritis, chorioamnionitis,
membrane premature
rup-ture, abortion, chronic
respiratory
diseasesincluding
pneumonia, and
meningitis.z'r
Ureaplasma
also
causesoccasionally respiratory
disease innewborn
andinfants.3
Deparfineûof Biology, Faculty
of
Medicine, Universityof
Indonesia, Jakarn, IndonesiaOne
of
the
factors
increasing
pathogenicity
of
urea-plasma
is
an
IgAl
protease. Apart from
Neisseria
gonorrhoeae,
N.
meningitidis,
Streptococcus
pneu-moniae,
S. oralis,
S. sanguis,
S. mitis,
Haemophilus
influe
lla
nocyt
pr
zyme
us
svery minute cells
and the easy lossof
its
viability
the characterization on theIgAl
protease ofureaplasma is
less advanced asin
the abovementioned bacteria. The
detection of ureaplasma colonies is
difficult
becauseof
their poor growth on
agar media.Lately,
development
of
PCR helped
to improve
clinical assessment.s'e
Finally,
the lack
of efficient in vivo
andin
vito
genetransfer
systems makesgenetic
studiesalmost
impos-sible.
The situation
will
improve
if
the
nucleotide
sequenceof the
genome
gets published.
To
be able142
MesakandSuhanaDKF3
and
used
for
comparison studies
also
U.urealyticumCXS.
We
wanted
to
characterize
the
ureaplasmal
IgAl
protease
activity
as
a
basis
for
enrichment
under-standing its
pathogenicity and
later-on
for
isolation
of
the
enzyme. Initial
steps
require cultivation,
main-tenance, and verificationof
ureaplasmacells. Then we
worked
out a
reliable
assay
for
the
IgAl
proteaseactivity based on results
from others.'
Proteaseactivity
of
human
IgAl
can
bevisualized by cleavage
of the
heavy chain
of
human
IgAl
as asubstrate by
SDSPAGE
and
Coomassieblue or silver staining.
Cells
were sonicated andfractionated
bysonication
or TritonX-100 or Triton
X-I14
extraction.
For further
charac-terization, we studied the kinetics
of
IgAl
proteaseactivity
andthe
localization of the protease
within the
ureaplasmacells.
Finally to analyze the presence
of
Fc
/
Fab
fragmentsafter digestion
of
IgAl
proteaseactivity
humanIgA1,
we also
developed semiquantitative
measurement
based on densityof
grayscales
andpixels
employing
Corel Photo
Paint
7
software.
The
resulted
scoreswere tabulated
on linear
graph
or
scatter diagram by
rsing Microsofi
Excel
97.METTIODS
Bacterial strains
U. ur e aly ti cum
Dl(F
3, U . ur e aly t ic umCX8
(was a gift
from J. Robertson)
and
Mycoplosma
pneumoniae
M129
(was
from the
stock collection
of
R. Herr-mann). Unlessotherwise
stated alway s U. urealyticum'DKF3 was used.
Media growth condition
and
maintenance
Two
hundred
microliters
(pl) of
deep-frozen
(-80oC)ureaplasma
cells
in
PBS
buffer
were thawed and
dilutèd
serially 1:10 up
to
l0-ewith
Bromothymol Blue
Broth, pH
6.0(2.l%oPPLO broth
without
crystal
violet
(Dif'co), 0.17o
least
extract, .0.004Vo
bromothymol
blue,
supplemented
with
lOVo
steril
normal
horse serumpH
6.0,
0.lVo
G}lL
solution
(Calbiochem)
and adjusteàpH
to
6.0).10
The
tubewith the
lowest
dilu-tion,
which
just
changedits color to
green,
asthe
pH
started to reach 6.2 assubcultured
for the
next 24 to 32hours
at
37oC.
st_oppedthe color
change
to
green
0-/, was
defined
asl0/
colorchang
encolordeveloped
in a transparent
optical view without
any turbidity.Med J Indones
Growth
onsolid
media was achievedby spotting
20pl
of
10-4, 10-5, and 10-6of
broth culture
intô
small Petri
dishes
containing Genital Agar
Media
(2Vo
PPLO
without
crystal
violet (Difco),
0.15Vo agar
no.l
(Oxoid), 0.lVo
yeast
extract,
l.l9Vo
HEPES buffer,
supplement
with
10 Vo sterilenormal horse serum
pH
6.0,
0.0257ourea, 0.l%o
G}JL solution
(Calbiochem)
and adjustpH
to
6.0;.ll
Mi"rou".ophilic
environment
was
simply
produced
by a
candle
flame, fading
after
several minutes
inside a tightly
closed
jar
containing
the
dishes.
Then
thejar
was incubated
for
3
days
at 37oC.Observation
wai made
using
a stereomicroscopewith
100-xmagnification.
Probe
designr
labeling, and
hybridization
to the
ureaplasma
genome.
A
twenty-two
-mer
oligonucleotide
as aspecific
probefor
the
ureaplasma species
was
designed based
onDNA
of urease sequencesfrom Genbank
databasewith
the accession numberof
X51315.
The sequenceof
theprobe
GAG
GTG
TAA
ACG GCT TAG TTA A.
was selectedby
employing OLIGO
4.0 software
(National
Biosciences
Inc.).
The
probenamed
Ul-ure
was
syn-thesized
by
DNA
Synthesizer
model 394A from
Ap-plied
Biosystems
at R. Frank's group
at
the ZMBH,
Heidelberg, Germany.
The oligo
was
purified
by
ethanolprecipitation
beforeit
waslabelled
at the 5' endby polynucleàtide
kinase ([y-32-P]ATP
).12Ureaolasma
DNA
isolation followed the method
of
SamËrook et al.t2
The genomic
DNA
was
digestedwith
restriction
endonuclease
EcoRI and the DNA
fragmens were separated
by
agarose
gel
electro-phoresis. Southern transfer
of
the chromosome
to
nylon
membrane
was later on
used
to
hybridize
theUl-ure.
The
experiments and
its
manipulation
were doneaccording to Sambrook
etat.t2
Auioradiography
was
developed
by
Phospholmager
(Molecular
Dynamics, Inc.).
IgAl
protease
activity assay,
time
course
of
the
assay,Triton
X-100 extraction
and Triton
X-114
phase
partitioning
Two
hundredsml
cultures
of
107color changing units
were
harvested
by
centrifugation and
washed
threetimes
with
PBSbuffer.
The
final
concentration
of
theproteins
was 20pgll in
aliquots
of
100pl.
As much
as300
pg of
total protein were mixed
with
3-5
pg of
humanIgAl
(Calbiochem).
As
apositive
control, I pg
of
pure
igase
from N. gonorrhoeae
was used,which
was
kindly
provided
by
S.C. Beck
of
theMax
Planck
Institût
ftr
Biologie,
Tubingen,
Germany.
As
anega-tive control, total cell protein of M.
pneumoniae was
used.
The
sample
mixtures
were incubated at
37oCovernight.
Sampleswere
boiled for
two minutes after
they had been
denaturedby protein lysis buffer
(125mM Tris-HCl pH
6.8,
4VoSDS,
l07o
p-mercapto-ethanol,
tÙVoglycerol,
and0.02Vobromophenol blue)
and
before
subjected
to SDS-PAGE.
The
kinetics
of
the
protease
activity
in
total
extracts
was
tested against thetime of incubation
athour
l,
3, 6.5, 10, 1 4.5,and
16.To
localize
the pro,teasein cellular fractions,
cells were sonicatedusing
BRANSON
815 cell
disrupter
bursts 8times
15 secondswith 10 seconds
interval. By
sub-sequent
centrifugation the
suspensionwas
separatedinto
sediment
and the supernatant.Triton X-100
extraction of total protein
extractyielded
a soluble
and
an insoluble
phase
asdescribeâ.I7 In
addition,
the
Triton
X-ll4
phasepartitioning
methodwas
ifications of
thepro-cedu
19
Fo.
this
purpose400
units
of
ureaplasmaculture
washarvested taken up
in
1ml,
washed threetimes
with
PBS,
andaliquoted
into
200pl of buffer
B(20mMTris
HCI pH
7.5,l50mMNaCl,
(1mM PMSF))
and
l%ov/v of Triton
X-114. It
wasmixed by
vortex-ing
and
then incubated at
4oC
overnight
with
mild
shaking and several
times re-vortexing.
Supernatantrvas
transferred
to
afresh tube
andincubated
at 37oCfor
30 minutes.
Subsequent steps
followed
asdescribed
by Proft
and
Henmann.le
The
Triton
X-114 phase
partitioning
and
the testing
for IgAl
protease
activity
was also donewith
frozen
(-80oC)
upto
threemonths ureaplasmal
cells.Bands analysis
using density
of
grayscales and
pixels
Protein
bands
in
SDS-PAGE
were
scanned
and analyzed byusing
density of grayscales andpixels
with
Corel
Photo
Paint
7software.
The
scannedpictures
were changed
to
grayscale (8 bit),starting
from
value 0 orblack
until
255
orwhite.
Thesoftware
can locate bandsaccording their
grayscaledensity in conjunction
with
thenumber
of
located
pixels. Level
equalization
was setautomatically.
Theruler
was also set to inpixel
units
with tick division
l0 per
tick.
Magic
wand masktool
was setinto normal with color
tolerance
modeof
lO (<
80Vo-
85Voof
255)or
3(>
80-
85Voof
255)
andused
to
locate bandsaccording
to their contours.
The automatichistogram
showed mean, median, and stand-arddeviation
of
thenumber
of pixels.
Pixels
in
local-ized
bands can
be
seenat
600x
or
1600x
magnifica-tions.
Measurement was done threetimes
to representthe core,
medium,
and outer area of bands. The average wascalculated
asvalue
of
the band
asdescribing
thescore,
which was obtained
by
multiplication
of
anaverage
of
(255-mean),
pixels
average
and
normal
value
used.
These
semi-quantitative
scores
can
beapplied
only
to a
distinct
band
of
the electrophoretic
result. Linear
graphsor
scatterdiagrams
of
the
scoreagainst
the test
parameters
were
done
by Microsoft
Excel 97 software.
RESULTS
After inoculation
of a
fresh
medium
atthe
ratio l:10
with
agrowing
ureaplamaculture
which
just
changescolor from
yellow
to green cells,could
bereproducibly
propagated
to 10/
color
changing
unit within
48 hoursat
37oC.
The
cultivation on solid
agar revealed
theformation
ofcolonies afterT2
hoursin
microaerophilic
environment
at37oC.
Colonies were very minute
but
distinctively differed from black teint
of
debris.
Thecolony has dark brown round
spots
on
the yellow
background
of
the
agarmedia. This
observation
wasconsistent
with
Robertson's description.l0'l
IAs
anadditional diagnostic
tool,
thegenomic
DNA
of
cultured
ureaplasma
cells
were probed
with
the
ureaplasma
specific oligo Ul-ure in
Southernblot
ex-periments
(Fig.
1).
As
canbe
seenfrom
lane
4
and 5from
Fig.lB
only
the
ureaplasma
DNA
cross-hybridized
with
the specific
probe
Ul-ure
it
was
present
although
atlowerconcentration
than thenega-tive control (Fig.
lA,
lane
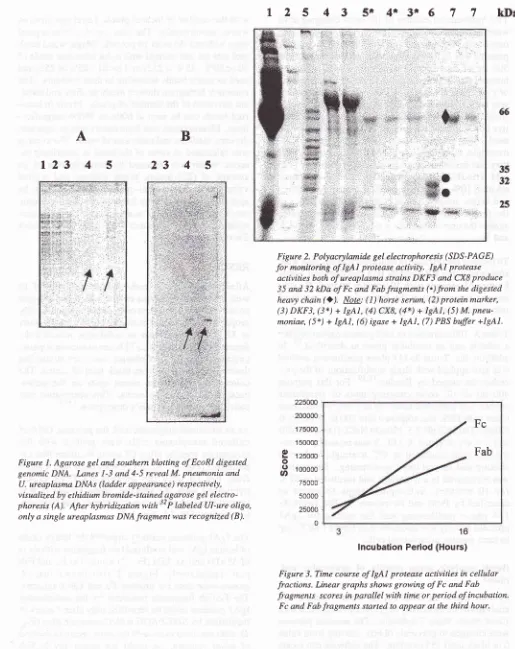
1-3).The
IgAl
proteasepartially
digested the heavy
chainof
humanIgAl
and producedtwo fragments
with
sizesof
35kDa and32 kDa (Fig.
2)
denoted asFc
and Fabpart, respectively. Figure
2
also shows
that
M.
pneumoniae
doesnot produce
Fc
andFab fragments.
The
FcÆab fragments produced
by
the
ureaplasmalIgAl
proteasecould
beidentified only
after
3 hoursof
incubation
by
SDS-PAGE
with
Coomassieblue (Fig.
3),
after onehour
evenwith
themost sensitive method
of
silver staining,
we
could
not
detect any
FcÆabi44
MesakandSuhanaA
123 4
5
B
23 4
5
Med J Indones
l2 5 4 3 51 4* 3*6
77
kDa
ffi
35 32
[image:4.595.67.582.73.722.2] [image:4.595.324.581.75.671.2] [image:4.595.346.578.337.622.2]25
Figure 1. Agarose gel and southern blotting of EcoRI digested genomic DNA.
Innes
1-3 and 4-5 reveal M. pneumonia andU. ureaplasma DNAs (Iadder appearance) respectively,
visualized by ethidium bromide-stained agarose gel electro-phoresis (A). Afrer hybridization with
"'P
labeled Ul-ure oligo,only a single ureaplasmas DNA fragment was recognized (B).
Figure 2. Polyacrylamide gel electrophoresis (SDS-PAGE)
for
monitoring ofIgAl
protease activity. IgAI protease activities both of ureaplasma strains DKF3 and CX8 produce 35 and 32 kDa of Fc and Fab fragments (.) from the digestedheavy chain
Q).
Npte: ( I ) horse serum, (2) protein marker, (3) DKF3, (3*) +IgAI,
(4) CX8, (4*) +IgAl,
(5) M. pneu-moniae, (5*) +IgAl,
(6) igase + IgA1, (7) PBS buffer +lgAI.Fc
Fab
16
Incubation
Period (Hours)Figure 3. Time course of IgAl protease activities in cellular fractions. Linear graphs shows growing of Fc and Fab fi'agments scores in parallel with time or period of incubation.
Fc and Fab fragments started to appear at the third hour.
225000 200000
1 75000 1 50000
I
rzsoooo
fi
rooooo75000 50000
To
analyzethe
localization
of
the proteaseactivity in
the
ureaplasmal cells
several methodswere applied to
produce
cellular
fractions.
The
phase separation wasdone
by
centrifugation
of
sonicated
cells
which
resulted
in
two fractions,
the pellet
and
the
super-natant.
Proteaseactivity
wasfound in
both fractions,
but
asjudged
by
signal
strength
in
SDS-PAGE,
thehigher activity
was
detectedin
the
supernatant.
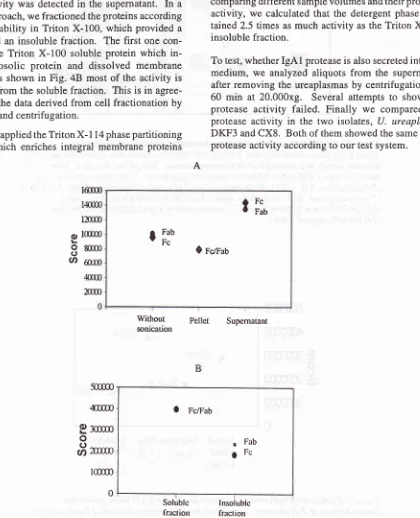
In
a second approach, wefractioned
theproteins according
to their solubility
in
Triton X-100, which provided
asoluble and
aninsoluble
fraction.
The
first
onecon-tains
all
the
Triton X-100 soluble protein which
in-cludes
cytosolic protein
and dissolved
membrane
protein.
As
shown
in Fig.
48
most
of the activity
is
recovered
from
the soluble
fraction.
This is
in
agree-ment
with
the
dataderived
from cell fractionation
by
sonication
andcentrifugation.
Finally,
we applied theTriton X- I
1 4 ph asepartitioning
method, which
enriches
integral
membrane proteins
and
can also
be
done
with
cells, which
were not
pretreated
by sonication.
The
fractionation
produced an aqueous phase, adetergent
phase,and
a detergentinsoluble
phase(Fig.
5).
The detergent
phaseshould
contain only the integral
membrane
proteins.
As
it
turned out, the
detergent phase showed
the
highest
activity (Fig. 6)
andthe insoluble the lowest
one. By
comparing different
samplevolumes
aridtheir
proteaseactivity, we
calculated
that the
detergent
phasecon-tained 2.5 times
asmuch
activity
as theTriton X-114
insoluble fraction.
To
test,whether
IgAl
proteaseis
also secretedinto
themedium, we
analyzed
aliquots from the
supernatantafter
removing the
ureaplasmasby centrifugation for
60
min
at
20.000xg.
Several
attempts
to
show
theprotease
activity
failed.
Finally
we
compared
theprotease
activity
in
the two isolates,
U.
ureaplasma
DKF3
andCX8.
Both
of
them showed the sameIgAl
proteaseactivity
according to our test
system.A
lm
o
l(m
88m
U)
oumDM
2m
5m
zm
9:m
o
8m
lm
Withour
sorucauon Supematant
r
i:,
g
i:'
ô
rcrau
o
FcÆab.
FabOFC
Solublc
[image:5.595.127.547.171.689.2]lrnclion
Figure 4. Distiburton of
IgAl
protease activities in cellular fracrtons. Scatter diagrams of Fc/Fab scores derived from sonicated cells (A) and Triton X-100 extraction (B). IgAI protease activities achieve higher scores in soluble fraction of both treatments.lnsolublc
146 Mesak and Suhana Med J Indones
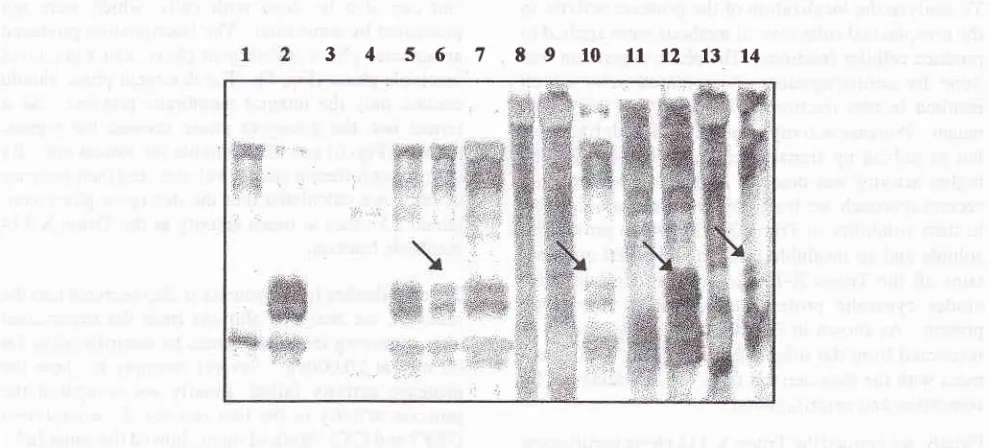
[image:6.595.84.579.85.309.2]34s6789tO11t21314
Figure 5. SDS-PAGEfor analysis of protease distibution
afterTritonX
extractions.Triton X-L14 phase partitioning and Triton X-100 uctraction showed that allfractions have
IgAI
protease activity, by producing Fc/Fab fragments (arrows). This gel was stained by silver.
Note:
(I)
IgAI
+ PBS
buffer, (2) ISAI + igase, (3) liquidphase -3
1tl, (4) liquidphase - 61t1,(5)liquidphase-51t1+lgAl,(6)detergentphase-21tL+lgAI,(7)detergentphase-51t|+lgAI,
(8)insolublephase-21t"1,(9)insolublephase-4ltl,(N)insolublephase-51t| +lgA1,
(lI)
solublefraction of TritonX-100, (12) solublefraction +IgAl,
(13) insolublefraction, (14) insolublefraction +IgAl.
a
FcÆabj
FcÆab1
FcÆabO
Fc/FLiquid
Detergent Phase InsolublePhase
(2
pt)
( 5 Ff)
Phase [image:6.595.211.459.394.664.2](spl)
(spt)
Figure 6. Distribution of
IgAl
protease activities after Triton X-[14 phase partitioning.Scatterdiagramof Fc/Fabscores derivedfromthelgAlproteaseactivitiesofliquid,detergent,
and insoluble phases of Triton X-| 14 partitioning. Soluble phase, which was pardrtoned into
DISCUSSION
The
clear green
color of
107color
changing
units
is
important to
produce
maximum
health
level
of
urea-plasmacultu.è.20
Cultiuuting
ureaplasmawithout
ureabut
with
thepH marker bromothymol blue
showed theimportance
of
the
color
changing
unit.
It
wasproven
that even
without
anycolor
change dueto
thelack of
urea in the
medium,
the cellsfrom
200ml culture could
be
harvested
between 16 and 24 hours
to
produce
2mg/ml of total cell
protein.
Moreover, culture medium
with
andwithout
ureagave the
samesignal intensity
in
the
bands
with
the
Fc
/
Fab fragments.
This
indicates that
IgAl
protease expressed
at the
samelevel during
thoseincubating
periods regardlessofthe
urea
supplementation.
Cultivation
onsolid
medianormallv
is donebv
subcul-turing
of
serial
dilution on
brotÉ .ulture.20'21
Al-though
it
needs special
skills
and
a
well-equipped
microbiology laboratory
to
assess ureaplasmal
colonies,
thebenefit of colony
verification
permits
thepossibility
to calculate colony-forming units.22
Be-sidesthat
thecombination
of liquid
andsolid
cultiva-tion
is a sensitive
methods,
which can be
done
for
diagnostic
purposes. But the
methods
of
molecular
biology
are
at
least as sensitive as cultivation
andcertainly much
faster
in
the
detection
of
ureaplasmacells.
Furthermore, the
Ul-ure
specific probe
wascross-reacting
with
the uraplasma genome but notwith
the
genome
of M.
pneumoniae,
a
non-urea
metabo-lizing mollicutes species.
Therefore, we believe that
this
Ul-ure oligonucleotide
can be used
as aspecific
probe
for
ureaplasmaswithin
thebacterial
classMol-licutes.
Digestion
of
thehinge region
of
theIgAl
heavychain
depends on
its interaction
with
thecatalytic
sitesof
theIgAl
proteaseat the
cell
surface
of
the
ureaplasmas.Based
on our
observation,
it
seemsthat
ureaplasmalcells
tend
to
aggregate andprecipitate
in
PBS
buffer.
This
waspreviously
assumedto
be the reasonfor
thepartial digestion
of theIgAl
heavychain. Butitturned
out that
this kind of
incomplete digestion
appearedin
both,
supernatant andpellet,
andsoluble
andinsoluble
fractions.
Hence,
it
is
presumed
that the activity
of
IgAl
proteasefrom
300
pg
of
total
ureaplasmal
cell
protein
used
is
less
than 1 pg pure
neisserial
igase.Nevertheless, one has also
to
consider
that
aprotein
extract might contain protease-inhibiting
substances and thatfurther enrichment
of the enzymewould result
higher
protease
activity.
A
reliable
assayto test
anIgAl
proteaseactivity
of
ureaplasma can be doneby
using the soluble part
of Triton X-100 extraction
andTriton X-114
phasepartitioning.
Fc
/
Fab
fragment
scoreswere higher in the
soluble
than
in
the insoluble
phases.
For
that
reason,
it
ispresumed
that the ureaplasmal
IgAl
protease
is
asoluble enzyme,
and theproteolysis
takesplace
in
thecytoplasm
or membrane-bound.
Since the
detergentphase,
which
contains theintegral proteins,
also has anactivity
against humanIgA1,
it
is mostprobable that
afraction of
this
enzymeis tightly
bound
to
the plasmamembrane
and
contains
a lipoprotein
moiety.
Bendjennat
et
a1.23showed
that
anuclease
from
M.
penetrans
has anactivity
both
in liquid
and detergentphases and
apparently
possesses alipoprotein
precur-sor component.
Meanwhile,
Washburn
et
al."*
reported that surface antigens
MAAl
andMAA2
ofM.
arthritidis,which
partition into
the detergent phase, areintegral
membrane-bound
proteins and
lipoproteins.
We
suggest that ureaplasmalIgAl
protease maturesin
cytoplasm before inserted
into
the
membrane
by
atransport
apparatuscontaining
protein-like
translocase(SecA
or SecY),
which was identified in
M.
pneu-moniae.25 The essential
secA
geneproduct recognizesthe
proteins
to
beexported
and catalysestheir
move-ment to the inner
cell
membrane.zÔNevertheless,
as analternative,
two forms of
ureaplasma
IgAl
proteasemay exist either membrane-bound or
cytosolic
with
thesame
hydrophilic
catalytic
site.
Onewould
expect thatthe protease
activity is
surface-exposedor
secretedto
prevent
IgAl
attack.4
So,if
the
proteasewould
be
acytosolic
enzyme,it
should beeffective
when thecells
are lysed.Acknowledgement
This work
waskindly
funded,
critically
reviewed
anddiscussed
by
Professor
R. Herrmann
of ZMBH,
andProfessor
H.-J. Freisleben
of
the
GraduateStudy
Pro-gram Biomedical
Sciencesof UI.
This
research
waspart of
Felix M.
Mesak's doctorate
work
supportedby
The
Excellent Student Scholarship
of
the
URGE
Project.
We
thank
Dr.
R.
Frank
for
synthesis
of
the
oligo-nucleotide,
and E.Pirkl
andD. Hofmann
for
introduc-ing the specific methods of
molecular biology
toFMM.
REFERENCES
l.
Taylor-Robinson D, GourlayRN.
GenusII.
Ureaplasma.sys-Mesak and Suhana
tematic bacteriology
Vol.
1.
Baltimore: The Williams andWilkins.
1984;770-52. Cassell GH, Waites KB, Watson HL, Crouse DT, Harasawa
R.
Ureaplasma urealyticum intrauterine infection: role inprematurity and disease
in
newboms. Clin Microbiol Rev1993;6:69-81
3. Tylor-Robinson. Infections due to species of Mycoplasma
atdUreaplasma: anupdate.
Clin
InfectDis
1996;23:671-84
4.
Kilian M,
Reinholdt J, Lomholt H, PoulsenK,
FrandsenEVG. Biological significance of
IgAl
proteases in bacterialcolonization and pathogenesis: critical evaluation
of
ex-perimental evidence. APMIS 1996 104:321-38
5. Kilian M, Brown MB, Brown TA, Freundt EA, Cassell GH.
Immunoglobulin A1 protease activity in strai ns of
Ureaplas-ma
urealyticum.
Acta Pathol Microbiol Immunol ScandSectB 1984;92:614
6. Robertson JA, Stemler ME, Stemke
GW.
ImmunoglobulinA
protease activityof
Ureaplnsma urealyticum.J Clin
Microbiol 1984; 19:255-8
7. Spooner RK, Russell WC, Thirkell
D.
Characterizationof
the immonoglobulin A protease of Ureaplasma urealyticum.
Infect Immun 1992; 6O:2544-6
8. Blanchard AJ, Hentschel J, Duffy L, Baldus K, Cassell GH.
Detection of Ureapalsma urealyticum by polymerase chain
reaction
in
the urogenital tract of adults, in amniotic fluid,and
in
the respiratory tractof
newborns. Clin Infect Dis1993; l7(Suppl):S148-53
9. Teng K,
Li
M, Yu W,Li
H, Shen D, LiuD.
Comparisonof
PCR with culture for detection of Ureaplasma urealyticum
in clinical samples from patients with urogenital infections.
J Clin Microbiol 1994; 32:2232-34
10. Robertson JA. Bromothymol blue broth: improved medium
for
detection
of
Ureaplasma urealyticurz(T-strain
mycoplasma). J Clin Microbiol 1978; 7:126-32
I
l.
Robertson JA, ChenMH.
Effectsof
manganese on thegrowth and morphology
of
Ureaplasma ureaLyticum.I Clin
Microbiol
1984:857-6412. Sambrook J, Fritsch EF, Maniatis
T.
Molecular cloning: alaboratory manual, 2nd ed. Cold Spring Harbor (NY): Cold
Spring Harbor Laboratory Press; 1989
13. Stoscheck
CM.
Quantitation of protein. Methods Enzymol 1990;182:50-55Med J Indones
Bradford
MM.
A rapid and sensitive method for thequan-titation of microgram quantities of protein utlliztng the
prin-ciple
of
protein-dye binding.Anal
Biochem 1976; 72:248-54
Laemmli
UK.
Cleavageof
structural proteins during theassembly of the head
of
bacteriophageT4.
Nature 1970;227:680-5
Morissey JH. Silver stain
for
proteinsin
polyacrylamidegels: a modified procedure
with
enhanced uniformsen-sitivity.
Anal Biochem 1981; 117:307-1017. Stevens MK, Krause
DC.
Mycoplasma pneumoniaecytad-herence phase-variable protein HMW3 is a component of the attachment organelle. J Bacteriol 1992; 17 4:4265 -1 4
18. Bordier
C.
Phase separation of integral membrane proteinsin Triton X- I 14 solution. J Biol Chem 198 I ; 256:1604-7
19. Proft T, Henmann
R.
Identification and characterization ofhitherto unknown Mycoplasma pneumoniae proteins. Mol
Microbiol 1994; 13:337 -48
20.
Thirkell D,
Myles
AD,
RussellWC.
Serotype8-
andserocluster-specific surface-expressed antigens of Ureap las
-rua urealy ticum. Infect Immun 1989 ; 57 :l 697 -17
0l
21. Stemke GW, Stemler ME, Robertson
JA.
Growthcharac-teristics
of
ureaplasmas from animal and human sources.Israel J Med Sci L984; 20:935-7
22. Runge M, Rykena S, Wildhagen K, Deicher H, Kirchoff H.
Detection
of
Ureaplasma urealyticumin
urine of patientswith systemic lupus erythematosus and healthy individuals
by culture and polymerase chain reaction. J Med Microbiol
1997;46:413-81
23. Bendjennat M, Blanchard A, Loutfi M, Montagnier L,
Bah-raoui
E.
Purification and characterization of Mycoplnsmapenetrans Caz*lMrg2* - dependent endonuclease. J Bacteriol
1997; 179:2210-20
24. Washburn LR, Weaver KE, Weaver
EJ,
Donelan W,Al-Sheboul
S. Molecular
characterizationof
l+lycoplasmaarthritidis variable surface protein
MAA2.
Infect Immun 1998;66:2576-8625. Himmelreich R, Hilbert H, Plagens
H, Pirkl
E,Li
B._C,Herrmann R. Complete sequence analysis of the genome of
the bacterium Mycoplama pneumoniae. Nucleic. Acid Res
1996;24:4420-49
26. Pugsley
AP.
The complete general secretory pathway inGram-negative bacteria. Microbiol Rev I 993 ;57:50- I 08
l5