Multiplex PCR for Rapid Detection of Rifampin and Isoniazid
Resistance
in Mycobacterium tuberculosis Isolated
from Bandung, Indonesia
HERA NOVIANA1,3*, ZEILY NURACHMAN1, MAELITA RAMDANI2,
AND AS NOER1
1Biochemistry Division, 2School of Biological Science, Institut Teknologi Bandung, Jalan Ganesha 10, Bandung 40132, Indonesia
3Microbiology Department, Medical Faculty, Universitas Katolik Atma Jaya, Jalan Pluit Raya No. 2, Jakarta 14440, Indonesia
Mycobacterium tuberculosis resistant to rifampin and isoniazid, known as multidrug-resistant M. tuberculosis (MDR-TB) strains, is an emerging problem of great importance to public health, with higher mortality rates than for drug-sensitive strains. Rifampin resistance is due to mutations on the hot spot region of the rpoB gene, especially at positions 526 and 531, and isoniazid resistance is due to mutation on katG at position 315. Mechanisms of resistance are an appropriate target for molecular genotyping diagnostic methods. Here we examined the multiplex PCR assays for the rapid detection targeting
rpoB526, rpoB531, and katG mutations. Sixty-one M. tuberculosis strains were studied based on rpoB526, rpoB531, and
katG315 assays employing multiplex PCR. Of the 61 strains, the susceptibility tests determined 42 isolates were MDR-TB strains, 10, 4, and 5 isolates were resistant to rifampin, isoniazid, and at least to six drugs which prescibed for TB, respectively. The mutation profiles of the 42 MDR strains assayed by multiplex PCR were 81 and 38.1% on rpoB and katG, respectively. Six rifampin-resistant isolates (60%) had a mutation on rpoB, 25% isoniazid-resistant isolates had mutation on katG, and 20% of the isolates that were sensitive to all drugs tested had a mutation on rpoB. Sequencing analysis revealed sensitivity of the multiplex PCR assay for rpoB was 98.4% and was 100% for katG. There was a 19% difference between phenotype and genotype properties of all isolates detected. In conclusion, the sensitivity of multiplex PCR method was sufficient for preliminary detection of rpoB and katG mutations, but resistance M. tuberculosis to rifampin and isoniazid were not always conferred by mutated alleles on rpoB and, especially, on katG.
Key words: multiplex PCR, rpoB, katG, sequencing
_____________________________________________
________________________
*Corresponding author, Phone: +62-21-6694366 ext. 254,
Fax: +62-21-6606123, E-mail: [email protected]
Mycobacterium tuberculosis, the causative agent of tuberculosis (TB), is still the single most deadly infectious disease in developing countries. The spread of MDR-TB, defined as M. tuberculosis resistant to at least isoniazid (INH) and rifampin (RIF), is worsening and emerging as a global emergency. It has increased worldwide and reached epidemic proportions in many countries (Fang et al. 1999; Rie et al. 2001; Wei et al. 2003). Indonesia is third in the rank of the highly burdened countries, after China and India. In 2001, the WHO estimates there are 122 TB patients per 100 000 people. The mortality rate is at least 140 000 people each year in Indonesia (WHO 2006).
RIF in combination with INH is a frontline anti-tuberculosis agent that is prescribed daily in millions of doses worldwide. Resistance to RIF is conferred by mutations in the rpoB hot spot region, encoding the subunit-b of RNA polymerase. The two most frequent mutations in the rpoB
are mutations at codon rpoB526 and rpoB531 (Gillespie 2002; Mokrousov et al. 2003; Van der Zanden et al. 2003; Werngren and Hoffner 2003). INH is a prodrug, which is peroxidatively activated by the M. tuberculosis catalase-peroxidase katG to produce damaging cell wall of the bacteria (Whitney and Wainberg 2002). Mutations at codon S315T (AGCaACC) were found in 60-70% of INH resistant strains (Van Doorn et al. 2001; Ramaswamy et al. 2003). This mutation was reported to be associated with intermediate or high levels of resistance to INH (1.0 to 10 mg l-1) (Mokrousov et al. 2002a; Caws et al. 2006). The inhA and kasA genes may participate in INH resistance.
Recent advances in the development of rapid and reliable diagnostic methods have allowed for detection of resistance to anti-tuberculosis drugs without the need for a viable culture. In this work we examined multiplex PCR assay as a means for rapid detection of rpoB526, rpoB531, and
katG315 mutation related to RIF and INH resistance,
respectively. The aim is to confirm results from molecular assays of multiplex PCR by nucleotides sequencing.
MATERIALS AND METHODS
Strains Collection. M. tuberculosis clinical isolates were recovered from different patients originating from Bandung, West Java with new or previous diagnosed pulmonary TB. Clinical specimens were collected from sputum, pleural exudate, and cerebrospinal fluid of patients between January and November 2005 at Balai Pengembangan dan Latihan Kesehatan (BPLK) laboratory for Mycobacteria, Department of Health and Rotinsulu Lung Diseases Hospital, Bandung, West Java.
Susceptibility Testing. Drug susceptibility was determined using the proportion method on Lowenstein-Jensen medium with the following critical concentration of the drugs: INH (0.2 mg 1-1), RIF (40 mg l-1), streptomycin (10 mgl-1), kanamycin (40 mgl-1), ethambutol (10 mg l-1), and pyrazinamide (40 mg l-1). The control was a tube of the medium without drugs inoculated by the strains of M.
tuberculosis being tested. All the tubes were incubated at
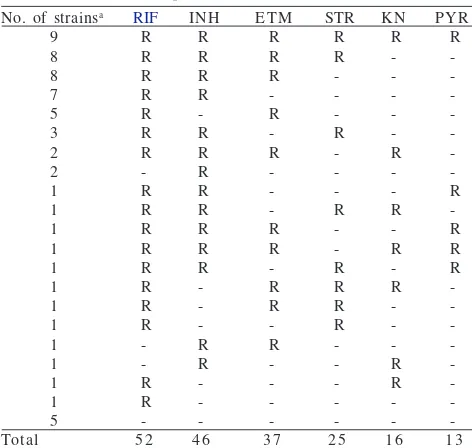
an = 6, RIF: rifampin, INH: isoniazid, ETM: ethambutol,
STR: streptomycin, KN: kanamycin, PYR: pyrazinamide. 9 Table 2 Resistance pattern of MDR M. tuberculosis strains iso
5 2 4 6 3 7 2 5 1 6 1 3
Table 1 Nucleotide sequences of primers used in the multiplex PCR assay and PCR for nucleotide sequencing
tube was ≤1% compared with bacterial growth in the respective control tube (Kumari and Ichhpujani 2000).
Preparation DNA Samples of M. tuberculosis. M.
tuberculosis colonies were lysed by 20 µl lysis buffer
(50 mM Tris-HCl pH 8.5; 1 mM EDTA pH 8.5; 0.5% Tween-20); and 4 ml Proteinase-K, using a 58 °C incubation temperature for an hour. The lysate was heated to 95 °C to disrupt Proteinase K, and the supernatant was then collected for further analysis. For both multiplex PCR and PCR for nucleotides sequencing, we used 5 ml DNA samples from this preparation.
Multiplex PCR. The multiplex PCR assay (modification of Mokrousov et al. 2002b; Mokrousov et al. 2003) was employed for mutational assay on rpoB526, rpoB531, and
katG315. Multiplex PCR, either for rpoB or katG, consisted
of three primers targeting codons rpoB526, rpoB531, and
katG315. A negative control was amplified using a standard
strain of M. tuberculosis, H37Rv. The primer sequences that used are shown in Table 1.
The inner forward primers are positioned so that their 3’-OH ends pair with the second bases of the respective codons in the wild-type allele. If a mutation occurs, this results in a mismatch at the 3’end of the inner primer sequence, in the absence of the variable PCR product (181 bp for rpoB526, 167 bp for rpoB51, and 292 bp for katG315). The invariable fragments of 249 and 435 bp are amplified by two outer primers, forward and reverse, flanked the rpoB hot spotand
katG region, respectively. The amplification a pair of forward and reverse primer is used to control the quality of the multiplex PCR. Meanwhile, the amplification employing similar forward and reverse primer without inner primer is used for nucleotide sequencing analysis.
The multiplex PCR reactions for katG315 were carried out in a thermal controller (Perkin-Elmer PCR system 2400, Inc.) under the following conditions: initial denaturation at 96 °C for 3 min; 30 cycles at 94 °C for 1 min, 58 °C for 40 s, and 72 °C for 30 s with the final elongation at 72 °C for 3 min. The PCR reactions for rpoB526 and rpoB531 were carried out under the following conditions: initial denaturation at 96 °C for 3 min; 5 cycles of 95 °C for 45 s, 60 °C for 1 min, and 72 °C for 30 s. Then 5 cycles at 95 °C for 40 s, 59 °C for 50 s, and 72 °C for 30 s. Then 25 cycles at 94 °C for 50 s, 55 °C for 40 s, and 70 °C for 30 s with the final elongation at 72 °C for 3 min. The PCR product was electrophoresed on 1.5% w/v agarose gel incorporating 0.5 g ml-1 ethidium bromide for visualization using a UV transilluminator.
RESULTS
Sixty-one strains of M. tuberculosis were collected from two laboratories and analyzed for their phenotype and
genotype properties. The susceptibility tests showed 42 strains were resistant to at least RIF and INH (MDR-TB), 10 strains were sensitive to INH but resistant at least to RIF, 4 strains were sensitive to RIF but resistant at least to INH, and 5 strains were considering sensitive to all of the drugs tested. The results of susceptibility test are shown in Table 2.
Molecular analysis of M. tuberculosis is a promising alternative to bacteriological assays. The genetic basis for RIF and INH resistance are mutations in the rpoB and katG genes, respectively. Using the method PCR-based, therefore the time to identify RIF and INH resistant strains were further is shortened to 2 h. In addition, it does not require any special probe preparation and can be applied to simultaneous analysis of several variable segments of the bacterial genome. Results of the multiplex PCR assays are shown in Fig 1 and 2.
Based on multiplex PCR results, this study determined the existance of various genotype combinations of M. tuberculosis strains as shown in Table 3.
DISCUSSION
There are many tools developed to simplify methods on TB diagnostics. The susceptibility test remains the gold standard method for phenotyping of M. tuberculosis. In total, drug-susceptibility testing of 61 M. tuberculosis isolates showed that resistance to RIF was the most frequent (85.2%) in this study, followed by INH resistance (75.4%). This study identified 42 strains of MDR-TB with nine of them (21.4%) resistant to six drugs tested. Resistance to ethambuthol, streptomycin, kanamycin, and pyrazinamide was observed for 60.7, 41.0, 26.2, and 21.3%, respectively.
Fig 1 Profiles generated by multiplex PCR targeting rpoB526
and rpoB31 (note: samples which amplified a product of 167 and 181 bp were considered to be wild-type rpoB531 and rpoB526, the strains with no amplified product of 167 and 181 bp were considered to be mutant, M: 100 bp marker lane).
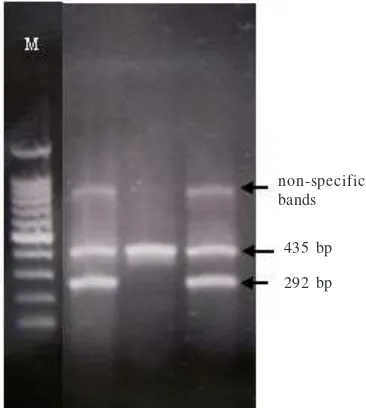
Fig 2 Profiles generated by multiplex PCR targeting katG315
(note: the strains with amplified product of 292-bp were considered to be wild-type, the strains with no amplified product of 292 bp were considered to be mutant, M: 100 bp marker lane).
non-specific bands
249 bp 167 bp
249 bp 181 bp
rpoB531 M rpoB526
rpoB531 M rpoB526
435 bp
292 bp non-specific bands
W: wild-type allele; T: mutant allele.
Table 3 Genotype combinations of MDR M. tuberculosis strains isolated from Bandung employing multiplex PCR
2 1 1 3 8 8 7 2 1 1
W W T W T W T T
W T W W T T W T
W W T T W T W T No. of strains
(n=61) katG315 rpoB531
Genotype
rpoB526
of MDR-TB has the capability to grow on media containing INH 10 µg ml-1 (data not shown). Mycobacterium tuberculosis strains carrying a mutated allele on rpoB531
had a high level resistance to RIF (MICe”100 mg l-1) compared with strains carrying a mutated allele on rpoB526. Likewise, M. tuberculosis strains carrying a mutated allele on katG315 had a high level resistance to INH (MIC between 1 mg l-1 and e”100 mg l-1) than strains that had no carrying mutated allele on katG315 (MIC 0.2-10 mg l-1).
Multiplex PCR assays for RIF and INH resistance tests rely on detection of mutations on rpoB526, rpoB531, and katG315. Of the 61 M. tuberculosis strains, approximately 68.9% were carrying mutated alleles of rpoB526 and rpoB531, whereas 27.9% carrying mutated allele of katG315. Mutation on rpoB526 and rpoB531 in this study were lower than reported earlier, with more than 95% of the rifampin-resistant M. tuberculosis strains caused by these mutations (Van der Zanden et al. 2003). Forty-two MDR-TB strains consisted of 16 (38.1%), 15 (35.7%), and 3 (7.1%) strains carrying the mutated allele of rpoB526, rpoB531, and both rpoB526 and rpoB531, respectively. Eight strains of MDR-TB were not detected carrying mutated alleles at either codon 526 or 531. Meanwhile, of the 42 MDR-TB strains analyzed by multiplex PCR for katG315, only 16 strains (38.1%) were detected carrying mutated allele of katG315. The results of genotype analysis on katG315 also were lower than reported in previous studies, which showed a 60-70% frequency for katG315 mutation. Mokrousov et al. (2002a) found a 93.6% prevalence of the katG315 mutation in strains from patients with both new and previous diagnosed cases of TB.
Of the ten strains sensitive to INH, but resistant to RIF when subjected to assay, seven were not demonstrated as having variable amplified product of 181 or 167 bp. This means that only three strains were detected carrying mutated alleles on rpoB526 and/or rpoB531. Interestingly, one strain sensitive to RIF but resistant to INH, and one strain sensitive to all six drugs, demonstrated had no amplified 181 bp product. Both strains that sensitive to RIF were considered to carrying mutated alleles on rpoB526. Those two strains might develop resistance to RIF at a low level (<40 mg l-1) so that in the susceptible test to RIF (40 mg l-1) they show as sensitive. Meanwhile, only one of four strains sensitive to RIF, but resistant to INH, was detected carrying a mutated allele on katG315.
There was a 500 and 1 000 bp of non-specific band in a few products of multiplex PCR for both rpoB526 and rpoB531, and katG315, respectively. The unspecific binding of primers or the abundant of DNA templates might cause this non-specific band (measurement of DNA samples were not done so that the concentration of DNA in every sample was different).
caused by inappropriate positioning of the PCR primers, which resulted in an underestimation of the prevalence of this mutation. Although Mokrousov et al. (2003) had been stably adjusted to assess reproducibility of multiplex PCR, it might be that the inner primer sequence and the effectiveness of the thermal controller was the crucial factor which affected the process of the multiplex PCR. The remaining isolates showed similarity between multiplex PCR and sequencing. This finding demonstrates that although the level of sensitivity of multiplex PCR to identify rpoB526, rpoB531, and katG315 mutations was not 100% because it was based on the specificity of multiplex PCR. However, this method can be used as preliminary assay for the detection of mutation on rpoB526, rpoB531, and katG315.
It is logical that a mutation would not be selected for in a population if it did not confer some form of selective advantage. The sequencing analysis recognized new mutated alleles in this study. Four of the eight MDR-TB strains carrying wild-type alleles on rpoB526, rpoB531, and katG315 had a mutation at a position both outside and within the hot spot of rpoB, either rpoB526 and rpoB531. It is remain unclear and undetermined what is the resistance mechanism of the remaining four MDR-TB strains. Interestingly, one strain sensitive to at least six anti-TB drugs demonstrated a new mutated allele carrying CCC (Pro) to CAC (His) at codon rpoB535, but the clinical significance of this mutation is not clear.
The high percentage of isolates lacking mutations suggests that phenotyping methods remain an important complement to genotyping methods for drug susceptibility testing. The genotype analysis determined 15 of the 52 (28.8%) strains which have phenotype resistant to RIF had no mutation on both rpoB526 and rpoB531. There were also 29 of the 46 (63%) strains phenotype resistant to INH which had no mutation on katG315. Therefore, this study revealed a 19% difference between phenotype and genotype properties of all isolates detected by multiplex PCR of rpoB526, rpoB531, and katG315. Our results suggest that this molecular method only determines the expected mutations, especially on rpoB526 and rpoB531. Susceptibility tests for the detection of INH resistance remain the most crucial assay because of the frequency of resistance corresponding to katG315 mutation being low. Therefore, although the molecular methods may aid in the rapid detection of mutations associated with drug resistance, the results always need to be confirmed by phenotyping methods.
In conclusion, the molecular analysis of M. tuberculosis is a promising alternative to bacteriological assays. The study demonstrates multiplex PCR is rapid, easy to perform, and sufficiently accurate for preliminary assay. It was essential that the test always be verified by standard culture-based methods because there were resistant strains of M. tuberculosis that were carrying no mutated alleles, whether on rpoB or katG. It is hope that the development of novel molecular methods using a rapid and reliable genetic approach will facilitate the appropriate and timely delivery of anti-tuberculosis therapy.
ACKNOWLEDGEMENT
We thank the technicians of the Balai Pengembangan dan Latihan Kesehatan, Public Health Province Laboratory
for Mycobacteria which contributed to the research. We
thank Francisca, Tintin, and Isak Solikin for assistance from their facilities and their advice and discussion. This study was supported by BPPS and Universitas Katolik Indonesia Atma Jaya.
REFERENCES
Caws M, Duy PM, Tho DQ, Lan NTN. 2006. Mutations prevalent among rifampin and isoniazid-resistant Mycobacterium tuberculosis isolates from a hospital in Vietnam. J Clin Microbiol
44:2333-2337.
Fang Z, Doig C, Rayner A, Kenna T, Watt B, Forbes KJ. 1999. Molecular evidence for heterogeneity of the multiple-drug-resistant Mycobacterium tuberculosis population in Scotland (1990-1997). J Clin Microbiol 37:998-1003.
Gillespie SH. 2002. Evolution of drug resistance in Mycobacterium
tuberculosis: clinical and molecular perspective. Antimicrob Agents
Chemother 46:267-274.
Kumari S, Ichhpujani RL. 2000. Guidelines on Standard Operating
Procedures for Microbiology. New Delhi: World Health
Organization Regional Office for South-East Asia. Module 31-32.
Mokrousov I, Narvskaya O, Otten T, Limeschenko E, Steklova L, Vyshnevskiy B. 2002a. High prevalence of KatG Ser315Thr substitution among isoniazid-resistant Mycobacterium tuberculosis clinical isolates from Northwestern Russia (1996-2001). Antimicrob Agents Chemother 46:1417-1424.
Mokrousov I, Otten T, Filipenko M, Vyazovaya A, Chrapov E, Limeschenko E, Steklova L, Vyshnevskiy B, Narvskaya O. 2002b. Detection of isoniazid-resistant Mycobacterium tuberculosis
strains by a multiplex allele-specific PCR assay targeting katG
codon 315 variation. J Clin Microbiol 40:2509-2512.
Mokrousov I, Otten T, Vyshnevskiy B, Narvskaya O. 2003. Allele-specific rpoB PCR assays for detection of rifampin-resistant
Mycobacterium tuberculosis in sputum smears. Antimicrob Agents
Chemother 47:2231-2235.
Ramaswamy SV, Reich R, Dou SJ, Jasperse L, Pan X, Wanger A, Quitugua T, Graviss EA. 2003. Single nucleotide polymorphisms in genes associated with isoniazid resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother 47:1241-1250. Rie AV, Warren R, Mshanga I, Jordaan AM, Spuy GD, Richardson M,
Simpson J, Gie RP, Enarson DA, Beyers N, Helden PD, Victor TC. 2001. Analysis for a limited number of gene codons can predict drug resistance of Mycobacterium tuberculosis in a high-incidence community. J Clin Microbiol 39:636-641.
Taniguchi H, Aramaki H, Nikaido Y, Mizuguchi Y, Nakamura M, Koga T, Yoshida S.1996. Rifampicin resistance and mutation of
the rpoB gene in Mycobacterium tuberculosis. FEMS Microbiol
Lett 144:103-108.
Van der Zanden AGM, Te Koppele-Vije EM, Bhanu VN, Van Soolingen D, Schouls LM. 2003. Use of DNA extracts from Ziehl-Neelsen-stained slides for molecular detection of rifampin resistance and spoligotyping of Mycobacterium tuberculosis. J Clin Microbiol
41:1101-1108.
Van Doorn HR, Kuijper EJ, Van der Ende A, Welten AGA, Van Soolongen D, De Haas PEW, Dankert J. 2001. The susceptibility of Mycobacterium tuberculosis to isoniazid and the Arg3Leu mutation at Codon 463 of katG are not associated. J Clin Microbiol
39:1591-1594.
Werngren J, Hoffner SE. 2003. Drug-susceptible Mycobacterium
tuberculosis Beijing genotype does not develop mutation
conffered resistance to rifampin at an elevated rate. J Clin
Microbiol 41:1520-1524.
Whitney JB, Wainberg MA. 2002. Isoniazid the frontline of resistance in Mycobacterium tuberculosis.J Med 6:114-123.