BY
CHARLIE DANNY HEATUBUN
THE GRADUATE SCHOOL
I, Charlie Danny Heatubun, certify that this dissertation is my own original work and has not been submitted in a previous application for a higher degree.
June, 25th 2009
and MIEN A. RIFAI.
Areca L. (Arecoideae: Areceae: Arecinae) is treated in this study as a genus of South Asian–West Pacific palm and best known for its type species Areca catechu, the betel nut palm. This genus comprises 41 species and included into two subgenera; subgenus Areca
and subgenus Beccarioareca. Seven species are newly recognized (A. bakeri Heatubun, A. churchii Heatubun, A. dransfieldii Heatubun, A. gurita Heatubun, A. mogeana Heatubun,
A. riparia Heatubun, and A. triginticollina Heatubun). Eight previously recognized species (A. jobiensis Becc., A. multifida Burret, A. nannospadix Burret, A. nigasolu Becc., A. rechingeriana Becc., A. rostrata Burret, A. torulo Becc. and A. warburgiana Becc.) are reduced to synonymy with A. macrocalyx Zipp. ex Blume; two species (A. guppyana Becc. and A. salomonensis Burret) are also reduced to A. novohibernica (Lauterb.) Becc. and one species (A. celebica Burret) to A. oxycarpa Miq.; and also one species A. macrocarpa
Becc. is also synonym to A. catechu L. While, three species (A. chaiana J. Dransf., A. congesta Becc., and A. ledermanniana Becc.) are considered to species incertae sedis (doubtful or uncertain species). A determination key is presented to subgenera and all easts of Wallace’s line taxa, including their detailed descriptions and for new species. A phylogenetic analysis of certain species in the genus was performed based on DNA sequences from two low-copy nuclear genes, phosphoribulokinase (PRK) and the second largest subunit of RNA polymerase II (RPB2). The monophyly of Areca as genus is reconfirmed together with its two lineages are also recovered. Subgenera and sections in the genus were assessed based on phylogenetic relationship together with their biogeography explanation. Natural history observations, including uses and conservation status are also presented in this study.
CHARLIE DANNY HEATUBUN. Sistematika dan Evolusi Marga Palem L.
Dibimbing oleh SRI S. TJITROSOEDIRDJO, JOHANIS P. MOGEA, WILLIAM J. BAKER dan MIEN A. RIFAI.
Areca L. (Arecoideae: Areceae: Arecinae) adalah marga palem yang distribusinya dari Asia Selatan sampai Pasifik Barat dan sangat terkenal dengan jenis tipenya Areca catechu, pinang sirih. Marga ini terdiri dari 41 jenis yang tergabung dalam dua anak marga; anak marga Areca dan anak marga Beccarioareca. Tujuh jenis adalah jenis baru (A. bakeri
Heatubun, A. churchii Heatubun, A. dransfieldii Heatubun, A. gurita Heatubun, A. mogeana Heatubun, A. riparia Heatubun, and A. triginticollina Heatubun). Delapan jenis yang sebelumnya dikenal (A. jobiensis Becc., A. multifida Burret, A. nannospadix Burret,
A. nigasolu Becc., A. rechingeriana Becc., A. rostrata Burret, A. torulo Becc. and A. warburgiana Becc.) merupakan sinonim baru untuk A. macrocalyx Zipp. ex Blume; dua jenis (A. guppyana Becc. and A. salomonensis Burret) juga merupakan sinonim A. novohibernica (Lauterb.) Becc. dan satu jenis (A. celebica Burret) untuk A. oxycarpa Miq.; demikian pula A. macrocarpa Becc. merupakan sinonim untuk A. catechu L. Sementera tiga jenis (A. chaiana J. Dransf., A. congesta Becc., and A. ledermanniana Becc.) dipertimbangkan sebagai jenis incertae sedis (meragukan atau jenis yang tidak pasti). Kunci determinasi ditampilkan untuk anak marga dan jenis-jenis yang berasal dari sebelah Timur Garis Wallace, demikian pula deskripsi lengkapnya dan juga untuk jenis baru. Analisis filogenetika untuk beberapa jenis pada marga didasarkan pada sekuens DNA dari dua low-copy gen inti; phosphoribulokinase (PRK) dan the second largest subunit of RNA polymerase II (RPB2). Monofiletik Areca sebagai marga kembali dikonfirmasi bersama dengan penemuan dua jalur evolusi di dalamnya. Anak marga dan seksi-seksi pada marga ini dikaji berdasarkan hubungan kekerabatan (filogeni) dan juga penjelasan tentang biogeografinya. Pengamatan tentang peri kehidupan alam termasuk kegunaan dan status konservasi juga ditampilkan dalam penelitian ini.
Corner, 1966; Moore & Dransfield, 1979). It is also the type genus of the family name
Arecaceae Bromhead. This genus is best known for its type species, Areca catechu – the widely cultivated betel nut palm, the seed of which is chewed with the leaf or inflorescence of Piper betle L. (Piperaceae), lime and sometimes tobacco and spices, principally as a mild stimulant. Betel nut is consumed by an estimated 200–400 million people world-wide (Gupta & Warnakulasuriya, 2002; Loo et al., 2006; Dransfield et al., 2008) – extensively in the Asia-Pacific region and expatriate Asian communities – making betel nut the fourth most widely “abused” substance after nicotine, alcohol and caffeine (Norton, 1998; Loo, et al. 2006).
The genus Areca has approximately 47 species (Dransfield et al., 2008), and is distributed from India and south China through Malesia to New Guinea and the Solomon Islands (Dransfield, 1984; Uhl & Dransfield, 1987; Dransfield et al., 2008).
The objective of this research is to provide a modern taxonomic treatment of Areca
including studies of its phylogenetic relationships, natural history, uses and conservation status. The work will be based on exhaustive studies of existing literature, a thorough examination of herbarium materials in international herbaria, extensive fieldwork and laboratory-based molecular systematic research. The project consists of four main subprojects: 1). Taxonomic revision of the genus Areca; 2). Species level phylogeny estimation; 3). Comparative morphological and molecular study of the genus Areca with aim to understand the morphological changes that occurred during their evolution; 4). and if possible to reconstruct the historical biogeography and origin of A. catechu L.
Taxon sampling and observations for the morphology and distribution of the species were based on herbarium samples or specimens (dried and spirit-preserved materials) deposited at international herbaria, namely A, AAU, B, BH, BO, BRI, FI, K, KEP, L, LAE, MAN, PNH, SAN, SAR, and SING (herbarium acronyms follow Holmgren et al., 1990), as well as the newly established small herbarium in Balai Penelitian Kehutanan
(Forestry Research Institute) in Manokwari, West Papua, Indonesia. Measurements were taken from spirit-preserved material and dried herbarium specimens and from living collections. Floral parts were measured from spirit-preserved material or dried specimens and rehydrated by boiling. Basic morphological characters such as habit, stem, leaves, inflorescence, staminate flower, pistillate flower, fruit, seed and their details were used to describe and recognize taxa; all morphological data was used for producing the descriptions of each taxon, while the key to species was constructed from the diagnostic characters only. The morphological species concept or taxonomic species concept was applied as a framework to define taxa, and assessed later with a phylogenetic species concept (Davies & Heywood, 1963; McDade, 1995; Gornall, 1997; Mayden, 1997; Dransfield, 1999), especially based on the result of molecular phylogenetics analysis of the genus Areca (Heatubun et al., in prep.). The conservation status of each species of the genus Areca in east of Wallace’s line was assessed based on the IUCN red list categories and criteria version 3.1 (IUCN, 2001).
outgroup (Norup et al., 2006; Dransfield et al., 2008; Baker et al., 2009). For DNA extraction, total genomic DNA extracted from silica gel-dried leaf materials (Chase & Hills, 1991) using the 2×CTAB method of Doyle & Doyle (1987). DNA were precipitated with 100% ethanol at –20°C, purified by cesium chloride/ethidium bromide gradients (1.55 g/mL) followed by dialysis and removal of ethidium bromide using butanol. Primer sequences for PRK and RPB2 were obtained from published sources (Lewis & Martinez, 2000; Lewis & Doyle, 2002; Roncal et al. 2005; Loo et al. 2006; Norup et al. 2006; Trenel
et al., 2007; Cuenca et al., 2007). Sequence alignment follow Loo et al. (2006) and Norup
et al. (2006) which every base position in the reverse and forward sequences were check and assembled using Sequencher 4.1 (Gene Codes Corp, Ann Arbor, Michigan, USA) and will be deposited in GenBank. The sequences then enter and aligned manually into data matrices in PAUP* version 4b10 (Swofford, 2002) for Macintosh computer. Phylogenetic analyses in this study were using parsimony analysis and Bayesian analysis. the standard procedures as follow: congruence between the PRK and RPB2 datasets will evaluated using the incongruence length difference (ILD) test of Farris et al. (1994) as implemented in PAUP* (Swofford 2002) and Mr. Test in MrBayes version 3.0b4 (Huelsenbeck & Ronquist, 2001). In parsimony analyses, uninformative characters and ambiguously aligned regions were excluded. All included characters were unordered and equally weighted. Initial analyses employed 1000 heuristic searches, each with starting trees obtained by random taxon addition, tree-bisection-reconnection (TBR) swapping, and keeping multiple trees per step (MulTrees on). Only groups that were found in the strict consensus tree and 50% or more of the replicates were recorded. Bayesian analyses were carried out using the program MrBayes version 3.0b4 (Huelsenbeck & Ronquist, 2001). Models of sequence evolution that the best fit the individual datasets were determined using Mr. Test. The models were evaluated by the Akaike information criterion (Akaike, 1973) implemented in the program. Parameters based on patterns in the data matrices. For all three datasets, four incrementally heated Markov chains were used in an analysis that was run for 100,000 generations initially with trees saved at every 10th generations. Trees produced prior to stationarity were discarded as the burn-in.
From this study, Areca is interesting palm genus not only for the species diversity but also for its natural history. This genus comprises 41 species and included into two subgenera; subgenus Areca and subgenus Beccarioareca. Seven species are newly recognized (A. bakeri Heatubun, A. churchii Heatubun, A. dransfieldii Heatubun, A. gurita
Heatubun, A. mogeana Heatubun, A. riparia Heatubun, and A. triginticollina Heatubun). Eight previously recognized species (A. jobiensis Becc., A. multifida Burret, A. nannospadix Burret, A. nigasolu Becc., A. rechingeriana Becc., A. rostrata Burret, A. torulo Becc. and A. warburgiana Becc.) are reduced to synonymy with A. macrocalyx
Zipp. ex Blume; two species (A. guppyana Becc. and A. salomonensis Burret) are also reduced to A. novohibernica (Lauterb.) Becc. and one species (A. celebica Burret) to A. oxycarpa Miq.; and also one species A. macrocarpa Becc. is also synonym to A. catechu L. While, three species (A. chaiana J. Dransf., A. congesta Becc., and A. ledermanniana
Becc.) are considered to species incertae sedis (doubtful or uncertain species). A determination key is presented to subgenera and all easts of Wallace’s line taxa, including their detailed descriptions and for new species.
Copyright © 2009, Bogor Agricultural University
Copyright are protected by law
1. It is prohibited to cite all or part of this dissertation without referring to and mentioning the source.
a. Citation only permitted for the sake of education, research, scientific writing, report writing, critical writing or reviewing scientific problems.
b. Citation does not inflict the name and honour of Bogor Agricultural University. 2. It is prohibited to republish and reproduce all or part of this dissertation without the
BY
CHARLIE DANNY HEATUBUN
G 361060021
As partial requirement fulfilment for the Doctoral Degree
in Plant Systematics
DEPARTMENT OF BIOLOGY
THE GRADUATE SCHOOL
BOGOR AGRICULTURAL UNIVERSITY
External Examiners
Examination I : Dr. Dra. Rugayah, M.Sc.
Herbarium Bogoriense, Puslitbang Biologi, Indonesia Institute of Sciences (LIPI), Cibinong, Bogor, Indonesia.
Dr. Dedy Duryadi Solihin, DEA
Head of Biology Study Programme, Faculty of Mathematics and Natural Science, Bogor Agricultural University,
Darmaga, Bogor, Indonesia.
Examination II : Dr. Timothy M. A. Utteridge
South-East Asia Section, Herbarium, Royal Botanic Gardens Kew, Richmond, Surrey TW9 3AB, United Kingdom.
Dr. Kuswata Kartawinata
Study Programme : Biology
Sub Study Programme : Plant Taxonomy
Approved by 1. Advisory Committee
(Dr. Sri S. Tjitrosoedirdjo, M.Sc) (Prof. Dr. Johanis P. Mogea) (Chairman) (Member)
(William J. Baker, M.Sc, Ph.D, FLS) (Prof. Dr. Mien A. Rifai) (Member) (Member)
2. The Biology Study Programme 3. The Graduate Schools
(Dr. Ir. Dedy Duryadi Solihin, DEA) (Prof. Dr. Ir. Khairil A. Notodiputro, MS) (Head) (Dean)
Many individuals and institutions contributed to the success of this work and I am grateful to them all. I would like to thank my advisors Dr. Sri S. Tjitrosoedirdjo, Professor Johanis P. Mogea, Dr. William J. Baker, Professor Mien A. Rifai and Dr. John Dransfield, whose always guided my work and gave me countless and useful recommendations to finished this dissertation. Professor Edwino S. Fernando is gratefully acknowledged for sharing his expertise on the Philippine’s Areca. Drs. Felix Forest, James J. Clackson and Professor Mark W. Chase to let me in to the lab, introduced me to DNA and Molecular Systematics, and guided me with the palm molecular phylogeny in the Jodrell Laboratory of Royal Botanic Gardens Kew. Drs. Christine D. Bacon and Raymond Baker from the Lyon Arboretum provided me with DNA materials of Areca from their garden collections in Hawaii. Dr. Tom D. Evans sent his collection of a new species of Areca from Cambodia. Professors Anders Barfod, Finn Borschenius and Henrik Balsev to invite me, arranged my travel and allowed me working with Areca collections in University of Aarhus and Copenhagen. Drs. Piero Cuccuini and Chiara Nepi also arranged my visit to Florence and allowed me working with the type specimens from Beccari’s historical collections. Ms. Julia Anak Sang and Mr. Shahbuddin Moh. Shabki are thanked for assistance with permits and all things regarding my fieldtrips to Sarawak. Dr. George Argent is responsible to my visiting in Edinburgh and Dr. Jef Veldkamp for my Leiden trip. Dr. Benito Tan and Ms. Serena Lee for accessing herbarium data base and type specimens of Areca kept in the Singapore Botanic Garden. I am grateful to the Keepers and/or curators of herbaria A, AAU, B, BH, BO, BRI, FI, K, KEP, L, LAE, MAN, PNH, SAN, SAR, SING and BPK Manokwari for access to their specimens, data bases, and loan materials for study. I also thank to Lucy T. Smith for preparing the wonderful plates for my new species. Drs. Rugayah, Dedy Duryadi Solihin, Timothy M. A. Utteridge and Kuswata Kartawinata are acknowledged for their willingness being examiners in my final examination.
Hermus Indou, Herkilaus Rumaikewi, Tobias Paiki, Maikel Simbiak and Piter Matani supported my research in various way. My colleagues, friends and my Papuan families in Bogor always offered good suggestion and words of hope during the hard times of my study.
Rector and the Dean of Fakultas Kehutanan Universitas Papua (UNIPA) Manokwari, Rector and the Dean of Sekolah Pascasarjana Institut Pertanian Bogor (IPB), they allow me to do my PhD. Financial supports came from BPPS Dikti Depdiknas and Royal Botanic Gardens Kew, UK for the PhD scholarships. The Royal Botanic Gardens Kew and Royal Botanic Gardens Edinburgh were funded my fieldtrips to Tanah Papua and North Sulawesi. My fieldtrips to Sarawak was granted by International Palm Society (IPS Endowment Fund 2007); visit to Nationaal Herbarium of Nederland, Leiden Branch by Flora Malesiana Foundation (Kostermann Funds) and Beasiswa Unggulan Dikti Depdiknas; visit to Royal Botanic Gardens Edinburgh, Scotland by the Sibbald Trust; travel to herbarium of University Aarhus and Monocot IV Symposium in Copenhagen, Denmark by the university of Aarhus; visit to Xishuangbanna Tropical Botanical Garden, Yunnan, China by Centre for Tropical Forest Science, Harvard University and China Academy of Sciences; and visit to herbarium Sezione Botanico, Meseo di Storia Naturale, Università degli Studi di Firenze, Florence, Italy by Royal Botanic Gardens Kew. The plates were funded by Pacific Biological Foundation and the Royal Botanic Gardens Kew.
Charlie Danny Heatubun is a son of Clemens Yoseph Heatubun (†) and Selfina Endemina Hursepuny (†). He was born on December 6th, 1974 in Manokwari, West Papua. In 1985 he was graduated from primary school, and secondary school in 1988 and also high school 1991, all in Manokwari, West Papua. He was continued his study at Department of Forestry, Faculty of Agriculture, Cenderawasih University in Manokwari and graduated in 1997.
Since 1997, he has been enrolled as academic staff in Forestry Department, Faculty of Agriculture; Cenderawasih University (now is Faculty of Forestry, University of Papua). In 2006, at the same year after finished his Master degree (M.Si) on Plant Taxonomy in Biology Department, the Graduate Schools of Bogor Agricultural University (SPs–IPB); he was continued his study to doctoral level in the same department of the same university with the scholarships from BPPS Dikti Depdiknas and the Royal Botanic Gardens Kew, UK. He was married Oktarina S. Simanjuntak and has a son Edward Glorious Excelsa Heatubun and a daughter Narcissa Elegantia Heatubun.
LIST OF FIGURES... xviii
LIST OF APPENDIXES... xix
GENERAL INTRODUCTION Background... 1
Objectives... 2
Literature Cited... 2
PHYLOGENY OF ARECA (ARECACEAE) BASED ON DNA Abstract... 6
Introduction... 7
Materials and Methods... 8
Results... 12
PRK Analysis... 12
RPB2 Analysis... 13
Combined Analysis... 15
Discussion... 20
Phylogenetics Value of PRK and RPB2... 20
Morphology... 21
Systematics Implications... 22
Biogeography... 22
Acknoledgements... 24
Introduction... 31
Materials and Methods... 32
Taxonomic Treatment... 35
Genus Description... 35
Infrageneric Classification of the Genus Areca... 36
Key to Subgenus of Areca... 36
I. Subgenus Areca... 37
II. Subgenus Beccarioareca... 54
Excluded and Uncertain Names... 72
Acknowledgements... 81
Literature Cited... 82
A MONOGRAPH OF ARECA OF EAST OF WALLACE’S LINE Summary... 88
Introduction... 89
Materials and Methods... 92
Results and Discussion... 93
Morphology... 93
Habit... 93
Stems... 94
Leaves... 94
Indumentum... 96
Inflorescences... 96
Pollination, Seeds Predation and Seeds Dispersal... 102
Biogeography... 104
Conservation Status... 105
Uses... 105
Taxonomic Treatment... 106
Key to Species of Areca East of Wallace’s Line... 106
Species Description of Areca East of Wallace’s Line... 107
1. Areca catechu L... 107
2. Areca macrocalyx Zipp. ex Blume... 113
3. Areca mandacanii Heatubun... 121
4. Areca novohibernica (Lauterb.) Becc... 123
5. Areca oxycarpa Miq... 127
6. Areca vestiaria Giseke... 129
Doubful and Uncertain Species... 132
Acknowledgements... 132
References... 133
Appendix: List of Specimens Examined and Identified... 139
GENERAL DISCUSSION Taxonomic Revision of the Genus Areca... 142
Phylogeny of the Genus Areca... 143
Literature Cited... 144
datasets.….…...11
3.1 Comparison of previously recognised taxa and infrageneric classification in Areca
1.1 The 50% majority rule consensus tree of the Bayesian inference analysis of PRK
dataset...16
1.2 The 50% majority rule consensus tree of the Bayesian inference analysis of RPB2 dataset...17
1.3 The 50% majority rule consensus tree of the Bayesian inference analysis of combined (PRK and RPB2) dataset...18
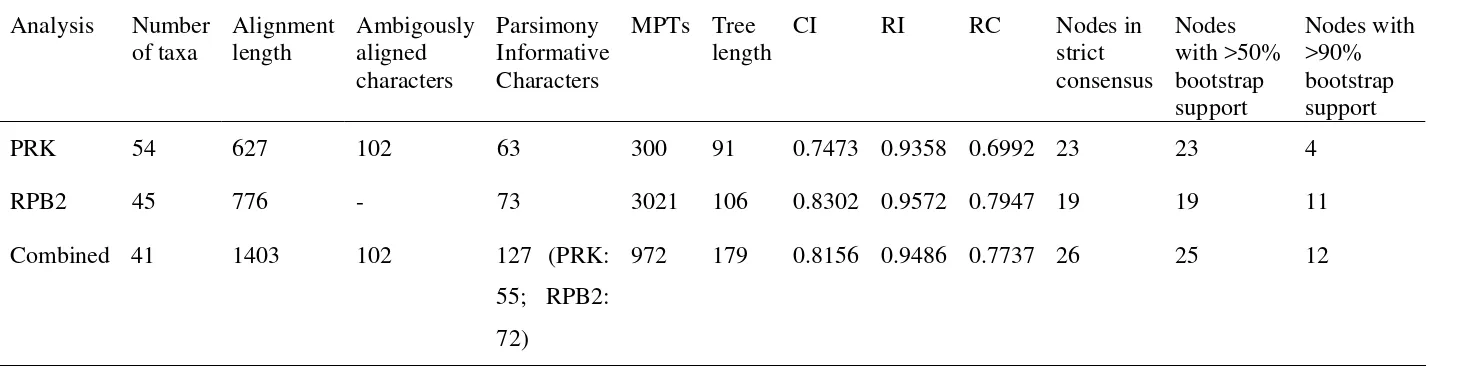
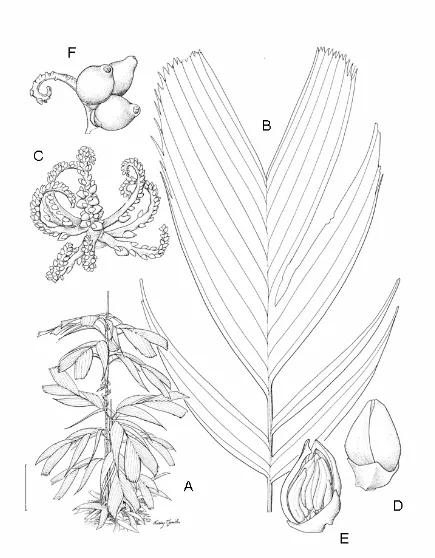
1.4 Areca bakeri Heatubun...40
1.5 Areca dransfieldii Heatubun...45
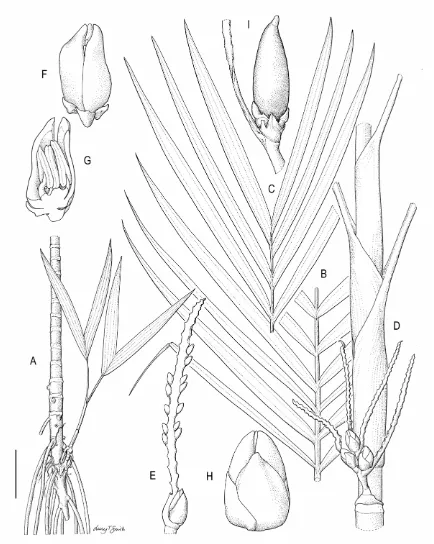
2.1 Areca riparia Heatubun...52
2.1 Areca churchii Heatubun...57
2.3 Areca gurita Heatubun...61
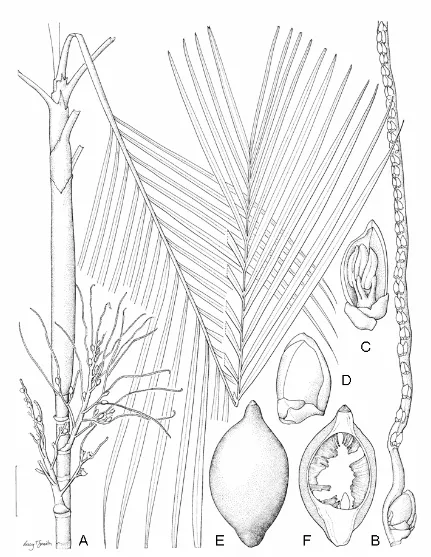
3.1 Areca mogeana Heatubun...66
3.2 Areca triginticollina Heatubun...70
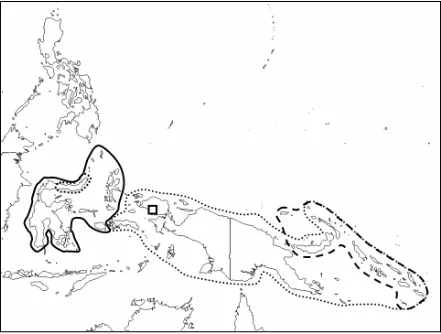
3.3 Distribustion map of the species Areca which native to east of Wallace’s line...91
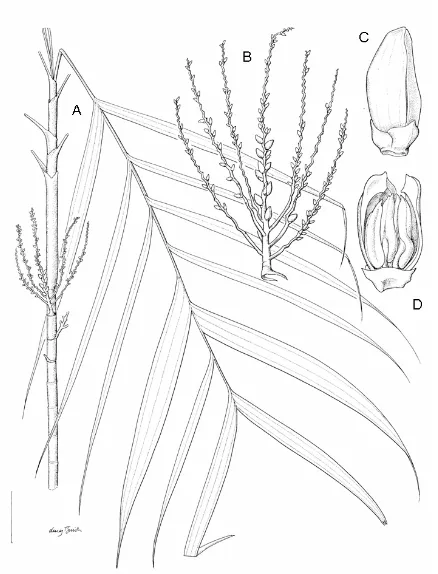
3.4. The inflorescence (infructescence) and the pistillate flowers morphology of Areca macrocalyx...97
3.5 Reproductive organ of Areca vestiaria and Areca oxycarpa...100
3.6 Areca macrocalyx Zipp. Ex Blume...116
general):
1. Dransfieldia (Arecaceae) – A New Palm Genus from Western New Guinea.
Systematic Botany 31: 61–69 (2006).
2. Apakah Jenis Itu? (Sebuah Pelajaran Penerapan Konsep Jenis Pada Suku Palem-Paleman Di New Guinea). Rampak Serantau 14: 190–199 (2007).
3. Two Species of Licuala (Arecaceae; Coryphoideae) from Western New Guinea.
Blumea 53: 429–434 (2008).
4. A new Areca from Western New Guinea. Palms52: 198–202 (2008)
Background
Areca L. was the first palm genus was described by Linneaus in Species Plantarum in
1753 based on Rumphius’ Pinanga published in Herbarium Amboinense in 1741 (see Corner,
1966; Moore & Dransfield, 1979). It is also the type genus of the family name Arecaceae
Bromhead. This genus is best known for its type species, Areca catechu – the widely
cultivated betel nut palm, the seed of which is chewed with the leaf or inflorescence of Piper
betle L. (Piperaceae), lime and sometimes tobacco and spices, principally as a mild stimulant.
Betel nut is consumed by an estimated 200–400 million people world-wide (Gupta &
Warnakulasuriya, 2002; Loo et al., 2006; Dransfield et al., 2008) – extensively in the
Asia-Pacific region and expatriate Asian communities – making betel nut the fourth most widely
“abused” substance after nicotine, alcohol and caffeine (Norton, 1998; Loo, et al. 2006). The
genus Areca also contains species of horticultural and agro-forestry significance (Ray &
Reddy, 2001) and recent work has revealed considerable potential for therapeutic drugs to be
extracted from some species, including cholesterol reducing agents (Byun et al., 2001) and
neurological and dermatological treatments (Lee & Choi, 1999; Sullivan et al., 2000).
The genus Areca has approximately 47 species (Dransfield et al., 2008), and is
distributed from India and south China through Malesia to New Guinea and the Solomon
Islands (Dransfield, 1984; Uhl & Dransfield, 1987; Dransfield et al., 2008). Although
varying in size from undergrowth palmlets to moderately tall tree palms, the genus is well
defined in its traditional concept by the presence of a crownshaft, infrafoliar inflorescences,
the single inflorescence bract (prophyll), complete floral triads confined to the basal part of
the branched inflorescence or its main axis, the symmetrical fruits with basally attached seed,
and the ruminate endosperm with basal embryo (Dransfield, 1984). However, Areca displays
perplexing variation in inflorescence structure and flower presentation that is in need of
further analysis, particularly in view of its wide distribution throughout Southeast Asia,
straddling Wallace’s line (Dransfield, 1995). In the recent phylogenetic classification of the
palm family, the genus Areca is placed together with Nenga and Pinanga in subtribe
Arecinae; tribe Areceae; subfamily Arecoideae (Dransfield et al., 2005, 2008; Loo et al.,
2006); differing from the previous classification which also included Gronophyllum, Gulubia,
Hydriastele,Siphokentia (these four now reduced to a single genus Hydriastele – see Baker &
The last infrageneric classification of the genus Areca was proposed by Furtado (1933)
comprising two subgenera and five sections: subgenus Blumeoareca (sections Arecella,
Oeotheanthe and Axonianthe); and subgenus Beccarioareca (sections Microareca and
Mischophloeus). However, these relationships within the genus are based on morphological
affinities alone (Furtado, 1933; Dransfield, 1984; Harley & Dransfield, 2003), and it has been
suggested that they need to be reassessed using modern methods (Dransfield, 1984).
Moreover, the geographic origin of economic species A. catechu is still uncertain, with the
Philippines (Beccari, 1919; Furtado, 1933), Malaysia (Corner, 1966; Jones, 1995) or Celebes
(Corner 1966) currently hypothesised to be the locality of the species’ origin.
Objectives
The general aim of this PhD project is to provide a modern taxonomic treatment of Areca
including studies of its phylogenetic relationships, natural history, uses and conservation
status. The work will be based on exhaustive studies of existing literature, a thorough
examination of herbarium materials in international herbaria, extensive fieldwork and
laboratory-based molecular systematic research. The project consists of four main
subprojects: 1). Taxonomic revision of the genus Areca; 2). Species level phylogeny
estimation; 3). Comparative morphological and molecular study of the genus Areca with aim
to understand the morphological changes that occurred during their evolution; 4). And if
possible to reconstruct the historical biogeography and origin of A. catechu L. The main
subprojects would be accommodating in the next following papers or chapters.
Literature cited
Asmussen, C. B. and M. W. Chase. 2001. Coding and non-coding plastid DNA in palm systematics. Amer. J. Bot.88: 1103−1117.
Baker, W. J. and A. H. B. Loo. 2004. A synopsis of the genus Hydriastele (Arecaceae). Kew Bull.59: 61−68.
Baker, WJ, Asmussen CB, Chase MW, Dransfield J, Forest F, Harley MM, Savolainen V, Uhl NW Wilkinson M. 2009. Complete Generic Level Phylogenetic Analyses of
Palms (Arecaceae) with Comparisons of Supertree and Supermatrix Approaches. Syst.
Byun, S. J., H. S. Kim, S. M. Jeon, Y. B. Park, and S. M. Choi. 2001. Supplementation of Areca catechu L. extract alters triglyceride absorption and cholesterol metabolism in
rats. Ann. Nutri. Metabol.45: 279−284.
Corner, E. J. H. 1966. The natural history of palms. London: Weidenfeld and Nicolson.
Dransfield, J. 1984. The genus Areca (Palmae: Arecoideae) in Borneo. Kew Bull.39: 1−22.
Dransfield, J. and N.W. Uhl. 1986. An outline of classification of palms. Principes 30:
3−11.
Dransfield, J. and N. W. Uhl. 1998. Palmae. In: Kubitzki, K (Ed.), the families and genera of vascular plants, vol. IV. pp. 306−389, Berlin: Springer.
Dransfield, J., N. W. Uhl, C. B. Asmussen, W. J. Baker, M. M. Harley and C. E. Lewis. 2005. A new phylogenetic classification of the palm family, Arecaceae. Kew Bull.60:
559−569.
Dransfield, J., N. W. Uhl, C. B. Asmussen-Lange, W. J. Baker, M. M. Harley and C. E. Lewis. 2008. Genera Palmarum: The Evolution and Classification of Palms. Kew: Royal Botanic Gardens Kew.
Flynn, T. 2004. Morphological variation and species limits in the genus Areca (Palmae) in New Guinea and the Solomon Islands. Unpublished Master thesis, University of Wales,
Bangor.
Furtado, F. X. 1933. The limits of the genus Areca L. and its sections. Repert. Spec. Nov. Regni Veg.33: 217−239.
Govaerts, R., and J. Dransfield. 2005. World checklist of palms. Kew: Royal Botanic Gardens Kew.
Gupta, P. C., and S. Warnakulasuriya. 2002. Global epidemiology of Areca nut usage. Addict. Biol.7: 77−83.
Hahn, W. J. 2002a. A molecular phylogenetic study of the Palmae (Arecaceae) based on atpB, rbcL and 18S nrDNA sequences. Syst. Biol.51: 92−112.
Hahn, W. J. 2002b. A phylogenetic analysis of the Arecoid line of palms based on plastid DNA sequence data. Mol. Phylogenet. Evol23: 189−204.
Harley, M. M. and J. Dransfield. 2003. Triporate pollen in the Arecaceae. Grana 41:
3−19.
Jones, D. L. 1995. Palms throughout the world. Sidney: Reed Books.
Loo, A. H. B., J. Dransfield, M. W. Chase and W. J. Baker. 2006. Low copy nuclear DNA, phylogeny and the evolution of dichogamy in the betel nut palms and their
relatives (Arecinae; Arecaceae). Mol. Phylogenet. Evol.39: 598−618.
Moore H. E., Jr. and J. Dransfield. 1979. Typification of Linnean palms. Taxon 28:
59−70.
Norton, S. A. 1998. Betel: consumption and consequences. J. Am. Acad. Dermatol. 38:
81−88.
Ray, A. K., and D. V. S. Reddy. 2001. Performance of Areca-based high density multi species cropping system under different level of fertilizer. Tropical Agriculture 78:
152−155.
Sullivan, R. J., J. S. Allen, C. Otto, J. Tiobech, K. Nero. 2000. Effects of chewing betel
nut (Areca catechu) on the symptoms of people with schizophrenia in Palau,
Micronesia. British Journal of Psychiatry177: 174−178.
Uhl, N. W. and J. Dransfield. 1987. Genera Palmarum: A Classification of Palms based on the work of Harold E. Moore Jr. Lawrance: L. H. Bailey Hortorium and International
Palm Society.
CHARLIE D.HEATUBUN1,2,3,4,WILLIAM J.BAKER3,JOHN DRANSFIELD3,JIM J.CLARKSON3, AND FELIX FOREST3
1
Fakultas Kehutanan, Universitas Papua, Jl. Gunung Salju, Amban, Manokwari 98314, Papua Barat, Indonesia;
2
Departemen Biologi, Sekolah Pascasarja Institut Pertanian Bogor, Kampus Darmaga, Bogor 16680, Jawa Barat, Indonesia;
3
Royal Botanic Gardens, Kew, Richmond, Surrey TW9 3AB, UK.
4
Author for correspondence (charlie_deheatboen@yahoo.com)
Areca is the type genus of the palm family Arecaceae and it distribute in the old world tropics from India, Srilanka, south of China to Malesian region to Solomon Island in the Pacific (Dransfield et al. 2008). The genus is well known from its type species, Areca catechu L., the betel nut palm or pinang. The betel nut palm is important commodity and has been developed to the large-scaled plantations for supplying the nuts demanding. Traditionally, the seeds are chewing for a mild stimulant (Loo et al., 2006; Dransfield et al., 2008), but in the recent years, the betel nut palm and other species of Areca have been used widely in therapeutic drugs; including cholesterol reducing agents (Byun et al., 2001), and neurogical and dermalogical treatments (Lee & Choi, 1999; Sullivan et al., 2000); horticulture and agroforestry significance (Ray & Reddy, 2001); and even further to maintain the biodiversity and conservation (The Economist, 8th November 2008: 100).
The genus Areca has approximately before 47 species by Dransfield et al. (2008) and reduces to 41 species in the recent revision (Heatubun et al., in prep.). Morphologically, the development of generic concept of Areca has been discussed in details by previous authors (Furtado, 1933; Dransfield, 1984; Uhl & Dransfield, 1987; Dransfield et al., 2008), including its relationships with other palm genera from morphology point of view (Harley & Dransfield, 2003; Baker et al., 2009).
In the recent phylogenetic classification of the palm family, the genus Areca is placed together with Nenga and Pinanga in subtribe Arecinae; tribe Areceae; subfamily Arecoideae
(Dransfield et al., 2005, 2008; Loo et al., 2006); differing from the previous classification which also included Gronophyllum, Gulubia, Hydriastele, Siphokentia (these four now reduced to a single genus Hydriastele – see Baker & Loo, 2004)and Loxococcus (Dransfield & Uhl, 1986, 1998; Uhl & Dransfield, 1987, 1999). While, the current phylogenetic evidence indicates that Areca is sister to a clade comprising Nenga and Pinanga (Loo et al., 2006; Norup et al., 2006; Baker et al., 2009).
The last infrageneric classification of the genus Areca was proposed by Furtado (1933) comprising two subgenera and five sections: subgenus Blumeoareca (sections Arecella,
Oeotheanthe and Axonianthe); and subgenus Beccarioareca (sections Microareca and
Mischophloeus). However, these relationships within the genus are based on morphological affinities alone (Furtado, 1933; Dransfield, 1984; Harley & Dransfield, 2003), and it has been suggested that they need to be reassessed using modern methods (Dransfield, 1984).
Moreover, the geographic origin of economic species A. catechu is still uncertain, with the Philippines (Beccari, 1919; Furtado, 1933), Malaysia (Corner, 1966; Jones, 1995) or Celebes (Corner, 1966) and western New Guinea (Heatubun, 2008) currently hypothesised to be the locality of the species’ origin.
The application of phylogenetics analysis to construct classification system in the palm family is widely accepted, and the sequences of specific DNA region also proved as powerful tools to solved taxonomic problem in palms. From DNA data used in the palms systematics, data from nuclear regions more variable and appear to evolve more rapidly (Dransfield et al., 2008), and this very useful at species level phylogeny analysis, especially low-copy nuclear gene (Lewis & Doyle, 2002). Recently, sequences of two low-copy nuclear genes have been shown to provide satisfactory phylogenetics information at lower taxonomic levels in the palm family (Bayton 2005; Lewis & Martinez 2000; Lewis et al. unpublished; Norup 2004; Roncal et al. 2005; Thomas et al. 2006), especially in subtribe Arecinae (Loo et al. 2006): nrDNA PRK (phosphoribolukinase, a Calvin cycle enzyme) intron 4 (Lewis & Doyle 2001, 2002) and RPB2 (RNA polymerase II) intron 23 (Roncal et al. 2005). And also have been successful in molecular dated phylogeny analysis to construct biogeography history and dispersal events in palms (Gunn, 2004; Norup et al., 2006; Trenel et al., 2007; Cuenca et al.,
2007).
Based on an existing preliminary dataset of subtribe Arecinae (Loo et al., 2006), it is already known that these regions are useful for resolving relationships among certain species in subtribe Arecinae, including Areca. Thus, sequences from PRK and RPB2 were used in this study; to evaluating monophyly and relationships within the genus Areca and testing subgeneric and sectional grouping proposed by Furtado (1933), and if possible to reconstruct biogeographic history and origin of the betel nut palm Areca catechu.
M
ATERIALS ANDM
ETHODSDNA-materials were collected directly in the fields by first author in western New Guinea (the Indonesian provinces of Papua and West Papua), North Sulawesi and Sarawak or obtained from botanical gardens, especially provided by Christine Bacon and Raymond Baker from Lyon Arboretum, Hawaii and Carl Lewis from Fairchild Tropical Botanical Garden Miami, Florida, USA. Also were using collections of DNA bank of Jodrell Laboratory, Royal
deposited in appropriate herbaria, and their identities confirmed.
All available sequences of species of the genus Areca were included in the taxon sampling, with species of Nenga and Pinanga (Loo et al. 2006), Bentinckia condapanna
(Arecoideae: Areceae: unplaced), Clinostigma savoryanum (Arecoideae: Areceae: unplaced),
Cyrtostachys renda (Arecoideae: Areceae: unplaced), Cyphokentia macrostachya
(Arecoideae: Areceae: Clinospermatinae) and Tectiphiala ferox (Arecoideae: Areceae:
Oncospermatinae) as outgroup (Norup et al., 2006; Dransfield et al., 2008; Baker et al., 2009).
For DNA extraction, total genomic DNA extracted from silica gel-dried leaf materials (Chase & Hills, 1991) using the 2×CTAB method of Doyle & Doyle (1987). DNA were precipitated with 100% ethanol at –20°C, purified by cesium chloride/ethidium bromide gradients (1.55 g/mL) followed by dialysis and removal of ethidium bromide using butanol.
Primer sequences for PRK and RPB2 were obtained from published sources (Lewis & Martinez, 2000; Lewis & Doyle, 2002; Roncal et al. 2005; Loo et al. 2006; Norup et al. 2006; Trenel et al., 2007; Cuenca et al., 2007). The primers for PRK (prk717f: 5’-GTG ATA TGG AAG AAC GTG G-3’, prk969r: 5’-ATT CCA GGG TAT GAG CAG C-3’) have been designed to be specific to the smaller of the two paralogues known in the palms (Loo et al., 2006) The primers for RPB2 (RPB2-PALM-INT23F: 5’-CAA CTT ATT GAG TGC ATC ATG G-3’, RPB2-PALM-INT23R: 5’-CCA CGC ATC TGA TAT CCA C-3’) are specific to palms. Each PCR product was amplified using a 25 µl reaction mix consisting of 1.1×ReddyMix PCR Master Mix (2.5mM MgCl2, ABgene), 0.3 µl of each primer (10mM,
final concentration), 1–5µl of template DNA, 0.9 µl bovine serum albumin (BSA) and optional dimethyl sulfoxide (DMSO) at 1% for recalcitrant amplifications of PRK and RPB2. Reaction mixtures will be subjected to the following temperature profile: initial denaturation at 95 °C for 5 min, 38 cycles of 96°C at 1 min each, 50°C for 1 min and 72°C for 1 min, and a final extension at 72 °C for 5 min. Amplification products were clean using the QIAquick PCR Purification Kit. The cleaned PCR products were cycle sequenced using the PRISM BigDye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems: ABI) following the manufacturer’s protocols. Amplification primers will use as sequencing primers. Subsequent cycle sequencing protocols and purification of cycle sequencing
10 products follow that of Asmussen and Chase (2001) and Salazar et al. (2003). The sequences were run on an automated sequencer (ABI).
Sequence alignment follow Loo et al. (2006) and Norup et al. (2006) which every base position in the reverse and forward sequences were check and assembled using Sequencher 4.1 (Gene Codes Corp, Ann Arbor, Michigan, USA) and will be deposited in GenBank. The sequences then enter and aligned manually into data matrices in PAUP* version 4b10 (Swofford, 2002) for Macintosh computer. Variable positions in the alignment will verified against the raw data to ensure that these will not a result of base-calling errors.
Phylogenetic analyses in this study were using parsimony analysis and Bayesian analysis. the standard procedures as follow: congruence between the PRK and RPB2 datasets will evaluated using the incongruence length difference (ILD) test of Farris et al. (1994) as implemented in PAUP* (Swofford 2002) and Mr. Test in MrBayes version 3.0b4 (Huelsenbeck & Ronquist, 2001). Character partitions were designated in the combined dataset and analyzed in 10000 replicates, each with a single heuristic search, saving trees with each replicate.
The two dataset partitions were analyzed separately and simultaneously. Taxon sampling varied between the PRK and RPB2 datasets, and therefore the combined dataset included only taxa for which both gene sequences were available.
For parsimony analyses, uninformative characters and ambiguously aligned regions were excluded. All included characters were unordered and equally weighted. Initial analyses employed 1000 heuristic searches, each with starting trees obtained by random taxon addition, tree-bisection-reconnection (TBR) swapping, and keeping multiple trees per step (MulTrees on). Only groups that were found in the strict consensus tree and 50% or more of the replicates were recorded.
11 Analysis Number
of taxa
Alignment length
Ambigously aligned characters
Parsimony Informative Characters
MPTs Tree length
CI RI RC Nodes in
strict consensus
Nodes with >50% bootstrap support
Nodes with >90% bootstrap support
PRK 54 627 102 63 300 91 0.7473 0.9358 0.6992 23 23 4
RPB2 45 776 - 73 3021 106 0.8302 0.9572 0.7947 19 19 11
Combined 41 1403 102 127 (PRK:
55; RPB2: 72)
R
ESULTSResults of the phylogenetics analysis of the genus Areca based on the molecular data nuclear DNA; PRK, RPB2 and combined as shown in the Table 1 and Figs 1–3.
PRK Analysis
In the PRK dataset, there were 54 sequences of 36 taxa, including 34 sequences from 20 species of Areca available. The average length of the genes is about 700 bp in the Areca
and the final length from data matrices is 627 bp and consisting of 23 bp exon 4, all of intron 4 and 130 bp exon 5, with 102 ambiguous characters and 63 parsimonious informative characters (Table 1). In the 5 samples, including A. chaiana was unsuccessful amplified and/or recovered apparently the existence of non-orthologous copies or pseudo-gene, thus all these genes were excluded from the analyses. Overall, parsimony and Bayesian inference analysis of PRK produced well-resolved and strongly supported topologies (Fig. 1). The parsimony bootstrap analysis resulted 300 the most parsimonious trees (MPT) with a length of 91 trees (CI = 0.7473, RI = 0.9538) and recovered 23 nodes over 50% bootstraps supports (one not shown in Fig. 1) including 4 nodes over 90% of bootstraps supported. As previously expected, that the Bayesian inference analysis will be generating more strong supported and more resolved in the PRK analysis than the bootstrap parsimony analysis. Of 25 resolved nodes in the Bayesian majority-rule tree, 21 nodes has value above 90% Bayesian posterior probability support and only 2 nodes below 75%.
The Arecinae is strongly resolved (100 Bayesian posterior probability, BPS and 90 bootstraps percentage, BP), whereas the outgroups is unresolved with polytomies (Bentinckia condapanna, Clinostigma savoryanum and Cyphokentia macrostachya) and poorly resolved in Cyrtostachys renda and Tectiphiala ferox (55 BPS and less than 50 BP). The topology supports the monophyly of Arecinae.
The monophyly of subtribe Arecinae which contain genera Areca, Nenga and Pinanga
is reconfirmed and Areca as sister to Nenga and Pinanga is resolved with strongly supports (100 BPS and 90 BP). And also Nenga is sister to Pinanga resolved with strongly supports in Bayesian (95 BPS) and moderate support in parsimony analysis (70 BP).
Areca is strongly supported (100 BPS, 91 BP) with two lineages to define two different clades. The discovery of two lineages within Areca with highly resolution (100 BPS in both, 89 and 55 BP) is indicate the infrageneric classification as proposed before by
are included in these two subgenera still need to re-arrangements based on the result of this study.
The subgenus Areca is strongly supports in Bayesian (100 BPS) and weak in parsimony analysis (55 BP). In general, the topology is low resolution with polytomies and the relationships among the species still unclear, except to A. triandra clade (A. triandra and
A. concinna) with strongly supports (100 BPS, 97 BP), the Philippines congested
inflorescence species clade (A. caliso, A. camarinensis and A. ipot) also with strongly supports (100 BPS, 96 BP) and A. oxycarpa (Heatubun 977 and Asmarayani 461).
The resolution in subgenus Beccarioareca is better than subgenus Areca, the node is highly resolved (100 BPS, 89 BP) and the relationships among the species in the subgenus more resolved. In this clade is also revealed that Areca furcata is sister to all species in subgenus Beccarioareca with relatively moderate supports (75 BPS, 69 BP).
The clade consist A. minuta, A. jugahpunya and A. ridleyana is resolved with high resolution in Bayesian analysis (100 BPS) and moderate in parsimony bootstrap analysis (71 BP). Also within the clade, the relationships between one accession A. minuta (Heatubun 892) and A. ridleyana recovered with strongly supports (100 BPS, 84 BP), whereas another accession of A. minuta (s.n.) and A. jugahpunya still unclear. While, two accession of A. minuta (Heatubun 887 and 888) are not clearly resolved as polytomies together with A. tunku.
Areca guppyana (= A. novohibernica) and A. vestiaria forming the Mischophloeus clade (94 BPS, 62 BP) and indicates A. novohibernica is sister to A. vestiaria. This clade is similar to section Mischophloeus of subgenus Beccarioareca in Furtado’s infrageneric classification (1933).
The other node is resolved with low supports (50 BPS, less than 50 BP) and expressed the relationships between A. insignis var. moorei and A. subacaulis. And this splits from other accession of A. insignis.
RPB2 Analysis
In RPB2 dataset, 54 sequences were in total from 36 taxa, including 30 sequences of 18 species of Areca were available. The length of the genes around 850 bp in the Areca and the final length from data matrices is only 776 bp of intron 23 with 73 parsimony informative characters. Similarly, about 7 samples unsuccessful, thus those sequences excluded from
RPB2 analysis and also in the combined analysis. The tree topologies in RPB2, nodes support and resolution performed by the parsimony bootstrap analysis lower than the results of Bayesian analysis. In the analysis produced 3021 most parsimonious trees (MPT) with a tree length 106 (CI = 0.8302, RI = 0.9572). Of 19 nodes resolved in majority rule tree 11 nodes over 90% BP (Fig. 2). Whereas in the Bayesian inference analysis, from 21 nodes resolved in the majority rule tree, 18 nodes exceeding 90% BPS.
The monophyly of Arecinae is reconfirmed with strongly supports in Bayesian analyisis (100 BPS) and moderate in parsimony bootstrap analysis (78 BP). From outgroups, only Clinostigma savoyarum and Cyrtostachys renda is resolved with strongly supports (100 BPS, 92 BP), while Bentinckia condapanna, Cyphokentia macrostachya and Tectiphiala ferox
are not clearly resolved their relationships.
Three lineages have been discovered from Arecinae in this analysis; two lineages (evolutionary lines) are also recovered in PRK analysis which contains Areca line and Nenga-Pinanga line and the third is Areca chaiana line. The topology is supports exclusion of Areca chaiana from Areca and as sister to all genera of Arecinae.
Areca is monophyletic and sister to Nenga and Pinanga are reconfirmed with strongly supports (100 BPS, 100 BP). Nenga is sister to Pinanga also reconfirmed from this analysis (90 BPS, 60 BP) and supports the monophyly of each genus in subtribe Arecinae.
Two lineages within Areca are also discovered with highly resolution in strongly supports in both analyses Bayesian and parsimony (100 BPS, 99 BP and 100 BPS, 100 BP). Two clades defined as clade of subgenus Areca and clade of Beccarioareca recovered with different resolution in tree topology. The relationships among species within subgenus in
Areca are more resolved than Beccarioareca.
Subgenus Areca is composed by nine resolved nodes with different resolutions. From those resolved nodes, three major clades have been identified as the congested inflorescence species clade, Areca catechu clade and Areca triandra clade. The congested inflorescence species clade is containing species accession of Areca from The Philippines, Sulawesi and New Guinea. This clade is strong supports in Bayesian analysis (95 BPS) and relatively weak supports in parsimony bootstrap analysis (62 BP), and from topology supports A. ipot as sister to all congested inflorescence species (A. camarinensis, A. caliso, A. macrocalyx and A. oxycarpa). Three accessions of Areca macrocalyx from New Guinea (Baker 1317, Heatubun 787 and 876) defined a resolved clade with strongly supports (100 BPS, 95 BP) and two of
than 50 BP). While, one accession of A. macrocalyx (Heatubun 798) is resolved separately from A. macrocalyx clade and nested together with Areca oxycarpa (Asmarayani 461 and
Heatubun 877) from north Sulawesi with strongly supports (100 BPS, 88 BP). The other clade is resolved with strongly supports (100 BPS, 87 BP) contain the Philippines species of
A. caliso and A. camarinensis.
Accessions of Areca catechu and A. mandacanii resolved in one clade with strong supports in Bayesian analysis (91 BPS) and unresolved in parsimony analysis (less than 50 BP). Although the relationships is not clear with polytomies especially between A. catechu
and A. mandacanii, except to two accessions of A. catechu (Heatubun 751 and 870) resolved (98 BPS, 62 BP), but this result supports Heatubun (2008) that A. mandacanii close to A. catechu based on morphology.
As in PRK analysis, A. triandra and A. concinna in RPB2 analysis also resolved in one clade with strongly supports (100 BPS, 96 BP). The relationships of among the accessions of A. triandra and A. concinna are not clearly resolved with polytomies.
The clade of subgenus Beccarioareca is low resolution in general and only A. minuta
(Heatubun 892) resolved as sister to all species in the clade with relatively moderate supports (76 BPS, 65 BP).
Combined Analysis
In the combined analyses, 41 sequences of 29 taxa, including 26 sequences of the 17 species of Areca with alignment 1403 bp long, including 102 ambiguous characters and 127 parsimony informative characters. In parsimony analysis, produced 972 most parsimonious tree (MPT) with a tree length 179 (CI = 0.8156, RI = 0.9846) in Table 1. Of 25 resolved nodes in majority-rule tree, 12 nodes over 90% bootstraps support (Fig. 3), and from 27 nodes on Bayesian inference analysis 23 nodes over 90%.
In general, the topology of tree in combined analysis is more resolved than previous analysis of each gene PRK and RPB2. Increasing the resolutions and supports in the tree topologies of the phylogenenetic tree can be carrying out with combine the PRK and RPB2. By combining these two genes in this analysis, is to maximize the informative characters (as phylogeny characters also) in both genes, so that the number of informative characters will increase and improve the topology of the phylogeny tree that produced.
FIG. 1. The 50% majority rule consensus tree of the Bayesian inference analysis of PRK dataset. Bayesian probability support values are shown above the brach, Parsimony bootstraps support values are shown below the branch. An asterisk (*) indicates a branch recovered with less than 50% bootstraps support. Infrageneric classifications within Areca (sensu Furtado, 1933) are indicated to the right of the species names.
FIG. 2. The 50% majority rule consensus tree of the Bayesian inference analysis of RPB2
dataset. Bayesian probability support values are shown above the brach, Parsimony bootstraps support values are shown below the branch. An asterisk (*) indicates a branch recovered with less than 50% bootstraps support. Infrageneric classifications within Areca (sensu Furtado, 1933) are indicated to the right of the species names.
FIG.3. The 50% majority rule consensus tree of the Bayesian inference analysis of combined (PRK and RPB2) dataset. Bayesian probability support values are shown above the brach, Parsimony bootstraps support values are shown below the branch. An asterisk (*) indicates a branch recovered with less than 50% bootstraps support. Geographical regions are indicated to the right of the species names and followed by Infrageneric classifications within Areca
(sensu Furtado, 1933).
to Nenga and Pinanga, the monophyly of Nenga and Pinanga and also Nenga is sister to
Pinanga are reconfirmed (Loo et al., 2006; Norup et al., 2006; Baker et al., 2009). Including resolved the relationships between two outgroups Clinostigma savoryanum and Cyrtostachys renda with relatively strong supports in Bayesian analysis (86 BPS) and moderate in parsimony analysis (65 BP), this result similar to Norup et al. (2006) and slightly different with Baker et al. (2009), which Cyrtostachys renda is sister to Clinostigma savoryanum and
Bentinckia condapanna. While, Bentinckia condapanna not resolved in this analysis.
Two lineages are discovered within Areca with strongly supports (100 BPS, 100 BP of each node) – sequence of A. chaina excluded from the combined analysis. Topologies are more clearly explained the relationships among the taxa in the clades, especially after incorporate with subgenera and sections sensu Furtado (1933) and their regions.
Subgenus Areca contains with three distinct clades and eight resolved nodes but A. dransfieldii and A. rheophytica still unresolved (less 50 BPS and BP). While, the clade of the congested inflorescence species of Areca from the Philippines, Sulawesi (Celebes), Moluccas, New Guinea and Solomon Islands resolved with strongly supports in Bayesian analysis (100 BPS) and moderate in parsimony analysis (76 BP). In this clade, is also discovered three evolutionary lines based on the geographic regions with highly resolution: the Philippines line (100 BPS, 81 BP) contains A. caliso, A. camarinensis and A. ipot; Moluccas-New Guinea-Solomon line (100 BPS, 98 BP) contains two accessions of A. macrocalyx (Baker 1317 and
Heatubun 876); and Sulawesi line (100 BPS, 95 BP) contains two accessions of A. oxycarpa
(Asmarayani 461 and Heatubun 877). And the Philippines line revealed that A. ipot is sister to A. caliso and A. camarinensis.
Areca catechu clade is resolved in high supports in Bayesian (81 BPS) but unresolved in parsimony bootstraps analysis (less than 50 BP). Similar to RPB2 analysis, the topology is low resolution with polytomies, except to two accessions of A. catechu from New Guinea (Heatubun 751 and 870) resolved (100 BPS, 60 BP). The relationships between all accessions of A. catechu and A. mandacanii are not clearly resolved however still indicated these two species are closely related (Heatubun, 2008).
Areca triandra clade is also resolved with strongly supports (100 BPS, 100 BP) including A. concinna in the clade. The relationships among the accession of A. triandra and
A. concinna is still not clearly resolved with polytomies.
Subgenus Beccarioareca is resolved with highly resolution (100 BPS, 100 BP) and A. furcata from Borneo is sister to all taxa in the clade. Two lineages also discovered (89 BPS, 65 BP); one nodes is resolved with relatively high supports in Bayesian (80 BPS) and collapsed in parsimony analysis (less than 50 BP), which forming a clade with contains A. guppyana (= A. novohibernica), A. subacaulis and accessions of A. vestiaria (Heatubun 879
and 885). The other node resolved with highly supports (100 BPS, 80 BP) and contains A. jugahpunya and two accessions of A. minuta (Heatubun 892 and s.n.).
Areca subacaulis from Borneo is resolved (96 BPS, 66 BP) as sister to “Mischopleous species” of A. guppyana (= A. novohibernica) from off-shore islands of Papua New Guinea (Manus Island, Bismarck Archipelago and Solomon Islands) and A. vestiaria from Sulawesi and North Moluccas. This result is never expecting before.
Areca guppyana (= A. novohibernica) is also resolved as sister to two accessions of A. vestiaria (Heatubun 879 and 885) with strong supports in Bayesian analysis (99 BP) and relatively moderate in parsimony bootstraps analysis (65 BP). The morphological characters also support this relationship (Furtado, 1933; Heatubun et al., inprep.).
While, the relationships among Bornean species of A. jugahpunya and two accessions of A. minuta still not clearly resolved with polytomies, although the clade is strongly supports.
D
ISCUSSIONPhylogenetic value of PRK and RPB2
The phylogenetic utility of the low-copy nuclear gene PRK and RPB2 have been proved useful in palm phylogeny (Lewis & Martinez 2000; Gunn, 2004; Norup 2004; Bayton 2005; Roncal et al. 2005; Loo et al., 2006; Norup et al., 2006; Thomas et al. 2006; Trenel et al., 2007; Cuenca et al., 2007; Baker et al., 2009). The level of variation in PRK and RPB2 is higher than other plastid genes used so far already discussed by Loo et al. (2006) and Norup
et al. (2006), including the comparison between number of parsimony informative characters obtained from the specific DNA regions, number of taxa sampled in the studies and the level of homoplasious characters gained from the analyses.
Despite from the limited number of samples used in study, percentages of parsimony-informative aligned positions for each partition in combined analysis (10.5% for PRK, 9.3% for RPB2) still exceeded from five plastid DNA regions used by (Hahn, 2002), which range
Asmussen & Chase, 2001). However, consistency and retention indices are high indicating low level of homoplasious characters in this study. And influence the resolution in phylogeny tree by providing the sufficient characters to resolve relationships among taxa of Areca and outgroups using on the study.
In this study has identified several new relationships and reconfirmed relations in the genus Areca addressed by morphology before (Furtado, 1933; Dransfield, 1984; Harley & Dransfield, 2003; Heatubun, 2008). We also discovered the compatibility of each region PRK and RPB2 to specific lineages in this palm genus, and by combining them, PRK and RPB2 will resolving the relationships in the lower taxonomic level such as species.
Morphology
In morphologically, these two lineages within Areca are supports by differences of the sepals of staminate flower and arrangement of the staminate flowers on the rachillae; the sepals being free and the staminate flowers uniseriate and distichous in subgenus Areca versus the sepal being united or calyx cup shaped and all or small portion of the staminate flowers spiral in subgenus Blumeoareca.
Whereas, in the sectional level is difficult to addressed, due to homoplasious morphological characters occurs. The morphological characters used to define sections in the previous classification (Furtado, 1933) have been evolved several times independently and separately in the genus Areca. For example, the small or even stem less habit was used to recognised the section Microareca in subgenus Beccarioareca (Furtado, 1933) also occurs in subgenus Areca – A. ahmadii (Dransfield, 1984) and in our result A. subacaulis nested together with A. guppyana (= A. novohibernica) and A. vestiaria of Furtado’s section
Mischopleous.
Areca chaiana in PRK analysis (unsuccessful sequence in RPB2, excluded in
combined) is sister to all genera of subtribe Arecinae, this taxon from morphological point of view is very distinctive and different from the generic concept of the genus Areca. Dransfield (1984) said that this is aberrant species of Areca when he was described it, because the nature of it inflorescence being spicate. More over, incomplete zonosulcate pollen aperture and tectum perforate reticulate are unusual in Areca were found in this taxon (Harley & Dransfield, 2003).
Systematics implications
Species of Areca that has been include to the subgenus and section from previous classification (Furtado, 1933; Dransfield, 1984; Harley & Dransfield, 2003; Heatubun, 2008) and applied to the phylogeny tree in combined analysis (Fig. 1–3). In general, this result is support infrageneric classification sensu Furtado (1933); two subgenera, namely Areca and
Beccarioareca, except to A. jugahpunya and A. tunku which were included in subgenus Areca
(Blumeoareca) section Arecella (Dransfield, 1984), whereas, in this result joins the clade defined as the Beccarioareca clade. Similar situation also found in sectional level within subgenera. In subgenus Areca, A. oxycarpa was in section Areca (Furtado, 1933) but in our results nested together with A. caliso, A. camarinensis, A. ipot and A. macrocalyx from section Axonianthe (Furtado, 1933). In previous infrageneric classification (Furtado, 1933),
A. concinna belongs to section Areca and nested together with A. triandra (section Arecella). Subgenus Beccarioareca has similar problem, which A. subacaulis (section Microareca, Dransfield, 1984) as sister to A. guppyana (= A. novohibernica) and A. vestiaria of section
Mischopleous (Furtado, 1933).
Although the limitation of samples used and unresolved branching nodes (polytomies) in topology of the phylogenetics tree, in fact, the clades grouping of our results are different from sectional grouping recognised in previous infrageneric classification (Furtado, 1933; Dransfield 1984; Harley & Dransfield, 2003), thus it will bring a taxonomic consequence, in which, we do not recommend to use the sectional classification within the genus Areca for the future application. We also strongly recommend excluding A. chaiana J. Dransf. from the genus Areca to proper palm genus.
Biogeography
From the topology of the phylogenetics tree (Fig. 3), suggests that the present distribution Areca is mainly caused by the long dispersal event rather than vicariance. And it might be, the dispersal events take place in several times dispersed out from centre of origin to recent area of distribution. Also from the tree topology, there is no indication of bimodal distribution pattern exist between west and east Wallace’s line in this genus and supports previous results from subtribe Arecinae (Loo et al., 2006). In our results, suggests Areca is
Borneo is sister to other species in the subgenus, including east of Wallace’s line species A. vestiaria and A. guppyana (= A. novohibernica). Confirmation of Borneo as the centre of origin for Areca is straight forward, however, Norup et al. (2006) was included Areca in Indian Ocean clade, and Loo et al. (2006) in the Sunda shelf as possible centre of origin from previous study in tribe Areceae and subtribe Arecinae. Of 20 from 41 species in the recent revision of the genus Areca (Heatubun et al., in prep.) are origins from Borneo and supports the idea of the centre of origin.
Loo et al. (2006) explained the diversification of the subtribe Arecinae (Areca, Nenga and Pinanga) is associated with the long, geologically stable history of the Sunda shelf and its ecological condition during Pliocene and Quaternary as proposed by Morley (1999). Furthermore, they also noted that palms were widespread in South-East Asia throughout much of Tertiary, and the subtribe Arecinae resembles the remnant of the early palm flora on that era. That evidence could be the explanation for distribution of A. triandra, this species distributed from India, Indochina, Thailand, Malay Peninsula, Sumatra, Java and Borneo.
Biogeographycal relationships between the west and east Wallace’s line taxa is clearly demonstrated in subgenus Beccarioareca. Explanations about how the species of Areca
crossed the Wallace’s line and reached islands of Sulawesi, the Philippines, New Guinea and the Solomon in the Pacific; as proposed by Hall (1998: 115); in the mid Oligocene, the southern-most promontory was the Sulawesi-Philippines-Halmahera arc which could have provided a pathway into the Pacific, via volcanic island stepping stone, for organisms that could cross sea water, and the other promontories terminated in the deep ocean area of the Pacific. Further molecular dated phylogeny analysis is needed to explain and/or reconstruct the dispersal events of Areca species in this region, including the diversification of the genus
Areca and the species Diaspora from the centre of origin in Borneo to westward and eastward cross the Wallace’s line, like similar study conducted by Gunn (2004) in Cocoeae with special emphasis on Cocos nucifera, Trenel et al. (2007) in the wax palm sub family Ceroxyloideae
and Cuenca et al. (2007) in tribe Chamaedoreeae.
Since the discovery of A. mandacanii (Heatubun, 2008), the country of origin of betel nut palm (A. catechu) become more and more blur – this also indicates in our results. As previously suggested, the betel nut palm might be come from the Philippines (Beccari, 1919; Furtado, 1933) and Malaysia (Corner, 1966; Jones, 1995) or Celebes (Corner 1966) and in
New Guinea by now. The suggestions above based observation on the wild species that morphologically close related to the betel nut. However, more samples are needed and must be include wild species from the Philippines in the analysis to try to solve the origin of betel nut.
A
CKNOWLEDGEMENTSMany individuals and institutions contributed to the completion of this paper. We would like to thank Drs Christine Bacon, Raymond Baker and Carl Lewis to provided DNA materials. The authority in Sarawak, especially the Sarawak Forestry Company (SFC) was allowed CDH to conduct his fieldtrip in Sarawak. Julia Sang and Shahabuddin M. Shabki are thanked for their helps with permit and all supports including organized CDH’s fieldtrip in Sarawak.
This paper is part of CDH PhD thesis which conducted at Institut Pertanian Bogor and Royal Botanical Gardens Kew. CDH would like to express his gratitude to Dr. Sri S. Tjitrosoedirdjo, Prof. Johanis P. Mogea, and Prof. Mien A. Rifai to their generous encouragements, supports, helps and guidance during the study. And the financial supports came from International Palm Society to funding CDH’s field trip to Sarawak through IPS Endowment Fund 2007. The Royal Botanic Gardens Kew–UK, BAT Biodiversity Partnerships and the Indonesian Ministry of Education (BPPS Dikti Diknas) to funding CDH’s PhD, to them are greatly acknowledged.
L
ITERATUREC
ITEDAsmussen, C. B. and M. W. Chase. 2001. Coding and non-coding plastid DNA in palm systematics. Amer. J. Bot. 88: 1103-1117.
Baker, W. J., C. B. Asmussen, M. W. Chase, J. Dransfield, F. Forest, M. M. Harley, V. Savolainen, N. W. Uhl, M. Wilkinson. 2009. Complete generic level phylogenetic analyses of palms (Arecaceae) with comparisons of Supertree and Supermatrix approaches. Systematic Biology. doi: 10.1093/sysbio/syp021