N
odulation – the symbiotic partnership between flowering plants and soil bacteria – is an ecologically and economi-cally important plant phenomenon. From an economic viewpoint, the ability of some plant groups to use atmospheric nitrogen, obviating the need for massive inputs of fertilizer, is a significant and desirable trait. A wealth of questions – ecological, developmental and molecular – are presented by nodulation. The process begins with signal exchange between bacteria and their plant hosts and leads to the production of a unique organ, the nod-ule, which contains novel components that are not found in either free-living bacteria or uninfected plants. From an evolutionary perspective, the factors leading to acquisition of this novel organ – a process that has apparently occurred several times independ-ently – are particularly fascinating. Phylogenetic studies have been undertaken with the aim of understanding relationships in both plants and bacteria, and of elucidating the evolution of individual genes and gene families within each group.Plant phylogenies: a predisposition to nodulate?
Nitrogen-fixing symbioses are best known from associations be-tween the legume family (Leguminosae or Fabaceae) and diverse Gram-negative soil bacteria (such as Rhizobium or
Bradyrhizo-bium; collectively ‘rhizobia’), in which nodules are produced on
plant roots or, in some cases, on stems. However, nitrogen-fixing symbioses in several other plant groups are also known.
Para-sponia, in the elm family (Ulmaceae), is the only one that is
nodu-lated by rhizobia. Cyanobacteria (for example, Anabaena and
Nostoc) form symbioses with plants as diverse as the water fern Azolla, cycads and the flowering plant Gunnera (Gunneraceae)1,2, and may even contribute to the nitrogen economy of cultivated grasses3. The Gram-positive actinomycete Frankia nodulates nearly 200 species of flowering plant belonging to eight families (Betu-laceae, Casuarinaceae, Coriariaceae, Datiscaceae, Elaeagnaceae, Myricaceae, Rhamnaceae and Rosaceae); taken together, actino-rhizal symbioses fix about as much nitrogen as the legume–rhizobia symbioses4,5
.
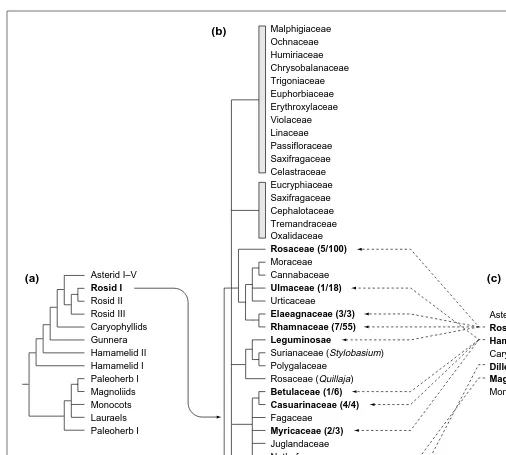
Flowering plant families involved in these symbioses have tra-ditionally been classified in different higher groupings, suggesting that symbioses evolved independently in diverse plant lineages. However, results from molecular phylogenetic studies have necessitated a dramatic revision of this view. Studies based on the chloroplast gene rbcL suggest that the flowering plant families involved in rhizobial or actinorhizal symbioses belong to the same large lineage5,6
(Fig. 1). This is interesting from a taxonomic per-spective, but what is more provocative about these results is the
possibility that a predisposition for forming a nodulation sym-biosis with rhizobial or actinorhizal bacteria has originated only once, in all of the hundreds of thousands of flowering plant species6
. The basis for such a predisposition remains a matter of speculation, with possible factors including unique classes of sec-ondary metabolites or phytoalexins; specific pathogens or patho-gen resistances; novel receptor molecules or hormone sensitivities; and unique physiological traits, such as nitrogen uptake or carbon metabolism4.
‘Predisposition’ and ‘origin’ are two quite different concepts, however. Within this ‘nitrogen-fixing clade’, nodulating genera are a small minority, and are scattered among non-nodulators, often even within the same family. This observation, along with the dif-ferences seen in nodule structure between legumes and actino-rhizal plants, and even among the actinoactino-rhizal families, suggests that nodulation has originated many times independently5
.
The origin(s) and evolution of nodulation in Leguminosae
Although as a group, legumes are well known for their nodulation symbiosis, not all genera nodulate. Most members of the sub-families Papilionoideae (which includes the typical ‘beans’) and Mimosoideae (which includes acacias and mimosas) appear to be capable of nodulating. By contrast, very few members of the third subfamily, Caesalpinioideae are known to nodulate7
. Based on the observation that rhizobia can infect roots of non-nodulating leg-umes, and that such plants show nitrogenase activity, it has been suggested that all legumes are capable of rhizobial symbiosis, although not all can form nodules8
. This speculative hypothesis could explain the high nitrogen content of the non-nodulating leg-umes9
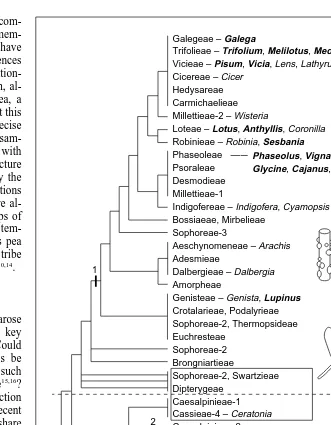
, but it leaves unanswered the question of the origin of nod-ules. A recent phylogenetic study, again using rbcL sequences, supports the long-standing hypothesis that the subfamily Caesal-pinioideae is a collection of diverse elements that includes the earliest-diverging lineages of the Leguminosae, and that most of the nodulating members of this subfamily belong to a lineage that includes Mimosoideae10
(Fig. 2). A simple parsimonious interpre-tation of the distribution of nodulation on the rbcL tree suggests as many as three independent origins of the syndrome: once in the mimosoid lineage, once in the only remaining confirmed caesal-pinoid nodulator, Chamaecrista, and a third time near the base of the Papilionoideae.
This hypothesis may be parsimonious, but is a phenomenon as complex as nodulation likely to have arisen independently so many times? Loss of nodules might be expected to occur more easily than gain, making hypotheses involving many independent
Phylogenetic perspectives on
nodulation: evolving views of plants
and symbiotic bacteria
Jeff J. Doyle
losses more plausible than those invoking even a small number of independent origins. Of course, the precedent already exists for multiple origins, in the phylogenetically diverse actinorhizal sym-bioses: as already noted, actinorhizal nodules in different fam-ilies are structurally diverse, as would be expected if nodules had different origins. Legume nodules are also developmentally and biochemically diverse, although they share many features that dis-tinguish them from actinorhizal nodules and even from the nodules of Parasponia. A phylogenetic assessment of the major nodule types in the Leguminosae10revealed that the unbranched, indeter-minate ‘caesalpinioid’ nodule type is scattered throughout the fam-ily and, notably, is present in Mimosoideae, Caesalpinioideae and
basal Papilionoideae (Fig. 2). This distribution is consistent with a single origin of nodulation in the family, with the caesalpinioid nodule type evolving in the common ancestor of all nodulating legumes.
Within Papilionoideae this basic caesalpinioid nodule type has been modified dramatically to produce the determinate ‘desmodi-oid’ nodules of many Phaseoleae, such as soybean (Glycine max) and the common bean (Phaseolus vulgaris), and the small, clus-tered ‘aeschynomenoid’ nodules found in the peanut (Arachis hypogaea) and its allies. Biochemical innovations have also oc-curred within the subfamily, and appear to be correlated with nod-ule type. For example, in the determinate nodnod-ules of Phaseoleae, Fig. 1. Phylogeny of nodulating flowering plants. (a,b) Phylogenies based on rbcL sequence data6
. Families that include nodulating genera are shown in bold, followed in parentheses by the number of nodulating genera and the total number of genera in the family. Details of relation-ships in exclusively non-nodulating lineages are not shown. (c) A traditional classification scheme for flowering plants. In the rbcL sequence phylogenies, all nodulating angiosperms and all symbiotically nitrogen-fixing angiosperms except Gunnera belong to a single extensive lineage, the Rosid I group, whereas in the more traditional scheme families involved in nodulation are scattered throughout most of the major subclasses. The unexpectedly close relationship revealed by rbcL suggests a predisposition to nodulation in this group of families6
. However, the scattered distribution of families and genera within the Rosid I lineage, along with structural differences in their nodules, is consistent with multiple origins of symbioses within this group5
.
Malphigiaceae Ochnaceae Humiriaceae Chrysobalanaceae Trigoniaceae Euphorbiaceae Erythroxylaceae Violaceae Linaceae Passifloraceae Saxifragaceae Celastraceae Eucryphiaceae Saxifragaceae Cephalotaceae Tremandraceae Oxalidaceae
Rosaceae (5/100)
Moraceae Cannabaceae
Ulmaceae (1/18)
Urticaceae
Elaeagnaceae (3/3) Rhamnaceae (7/55) Leguminosae
Surianaceae (Stylobasium) Polygalaceae
Rosaceae (Quillaja)
Betulaceae (1/6) Casuarinaceae (4/4)
Fagaceae
Myricaceae (2/3)
Juglandaceae Nothofagaceae
Coriariaceae (1/1)
Tetrameleaceae
Datiscaceae (1/3)
Cucurbitaceae Begoniaceae
Other 'Rosid I' dicots Asterid I–V
Rosid I
Rosid II Rosid III Caryophyllids Gunnera Hamamelid II Hamamelid I Paleoherb I Magnoliids Monocots Lauraels Paleoherb I
Asteridae
Rosidae Hamamelidae
Caryophyllidae
Dilleniidae Magnoliidae
Monocots
(a) (c)
nitrogen transport involves ureide com-pounds instead of the amides used by mem-bers of other tribes, most of which have indeterminate nodules. Such differences may also extend to the linkage relation-ships of genes involved in nodulation, al-though data are as yet scarce. In pea, a number of such genes are linked11, but this is not the case in soybean12
. More precise phylogenetic hypotheses for a wider sam-ple of papilionoid legumes, cousam-pled with more detailed surveys of nodule structure and chemistry, are needed to identify the points at which evolutionary modifications occurred. Phylogenetic hypotheses are al-ready being produced for some groups of papilionoid legumes, for example the tem-perate herbaceous clade that includes pea and alfalfa13
, and the Millettieae, a tribe with many connections to Phaseoleae10,14.
Developmental conservation and origins of legume nodules
Regardless of whether or not nodules arose one or more times in legumes, a key question is, how did they evolve? Could features of nitrogen-fixing symbiosis be derived from other, older symbioses, such as those involving endomycorrhizae15,16
? Both symbioses involve some induction of the host defense response15
, and recent studies suggest that both phenomena share a common signal transduction pathway17
. Mutualistic symbioses of both types could have arisen from still earlier pathogenic interactions15.
To what other structures are nodules hom-ologous in a developmental sense? This is an interesting question regardless of whether or not legume nodules all have a single origin (are homologous in a phylogenetic sense), or of the relationship between nodu-lation and other symbioses, and is irrespec-tive of the ability of non-nodulating legumes to derive benefit from rhizobia8
. However, it is not a simple question, and we lack sufficient information to answer it satisfac-torily15. By analogy, even if the flower is considered to be a condensed shoot bearing modified leaves, and is thus homologous to the shoot, it is still a novel organ. Knowl-edge of this developmental homology does
not explain the evolution of flowers. The legume nodule, although commonly produced on roots by rhizobial infection of root hairs, has a stem-like anatomy, in contrast to the anatomically more root-like nodules of actinorhizal plants18. The legume nodule is clearly not a stem, however, regardless of whether it might have been formed by recruitment of developmental programs normally associated with stems. Nodules contain what has been termed a ‘novel organelle’19, the symbiosome. This organelle consists of bacteroids (rhizobia in their modified, symbiotic phase) enclosed in a host peribacteroid membrane through which carbon and other nutrients are passed to the bacteria, and nitrogenous compounds are transferred to the host plant.
The presence of nodule-specific proteins, called nodulins, was also taken as evidence that the evolutionary novelty of nodules extended to the molecular level. However, it is likely that most nodulin genes are also expressed elsewhere in the plant and have been recruited from other functions20
. For example, uricase II was thought to be expressed only in the nodules of soybean, but is now known to be expressed in cotyledons too21
. Thus, one challenge in understanding the evolution of nodulation is to determine the ancestral expression patterns of recruited nodulin genes. Data on the expression of early nodulin (ENOD) genes in mycorrhizal symbioses17
are clearly of great interest, as are studies of genes essential for, and uniquely associated with, nodulation, but even
Fig. 2. Phylogeny of Leguminosae, as hypothesized from rbcL sequence data10
. All groups above the broken line belong to the largest subfamily, Papilionoideae; below the line are Caesalpinioideae and Mimosoideae. Tribal names are used, sometimes several times where tribes consist of unrelated lineages (e.g. Caesalpinieae and Sophoreae). Important or char-acteristic genera are listed for some tribes; those shown in bold also appear in Fig. 4. Groups in boxes are not known to nodulate. Possible independent origins of nodulation, as sug-gested by parsimonious optimization of this trait, are shown as numbered vertical bars along particular branches. Drawings of the major nodule types appear next to some of the groups in which they occur. Unbranched, elongate, indeterminate nodules are ‘caesalpinioid’; branched, indeterminate nodules are ‘astragaloid’; large, round, determinate nodules are ‘desmodioid’; small, clustered nodules are ‘aeschynomenoid’.
Galegeae – Galega
Trifolieae – Trifolium, Melilotus, Medicago
Vicieae – Pisum, Vicia, Lens, Lathyrus
Cicereae – Cicer Hedysareae Carmichaelieae Millettieae-2 – Wisteria
Loteae – Lotus, Anthyllis, Coronilla
Robinieae – Robinia, Sesbania Phaseoleae
Psoraleae Desmodieae Millettieae-1
Indigofereae – Indigofera, Cyamopsis Bossiaeae, Mirbelieae
Genisteae – Genista, Lupinus Crotalarieae, Podalyrieae
MIMOSOIDEAE – Leucaena, Acacia
the expression patterns of more prosaic ‘housekeeping’ genes could contribute to our understanding of nodulation and nodule homology. Independent origins of nodules would mean the inde-pendent recruitment of ‘nodulin genes’ from within a gene family. This could lead to the cognate nodulin genes of different legumes being paralogous (i.e. different duplicate copies or members of different subfamilies). By contrast, the assumption is that the com-mon ancestor of species possessing homologous nodules recruited a single gene family member as its nodulin22. If there have been no shifts in gene expression, the species that are derived from this ancestor should all use the same family member (orthologue) as their nodulin gene. A test for nodule homology thus involves developing rigorous phylogenetic hypotheses for numerous gene families that include nodule-expressed members.
The phylogeny of genes: nodulin family trees
Genes involved in nodulation often belong to multigene families, whose evolutionary patterns can be very complex as a result of nested duplications, gene losses and concerted evolution23. This complexity is well illustrated by genes encoding leghemoglobin. Although globins in plants were originally identified in the nodules of legumes (and were thus the quintessential nodulins), globin genes have now been found in the genomes of both non-nodulating legumes and also in non-legumes24
. In the nodule, the oxygen-binding ability of leghemoglobin contributes to maintain-ing an environment that simultaneously permits bacterial growth and restricts oxygen availability that would inhibit bacterial nitrog-enase. In other tissues, it is speculated that globins may serve other functions such as oxygen sensing25. The plant globin gene
tree is very complex, and it is clear that there is still much to be learned about this gene family. The observation of marked phylogenetic diversity of globin genes of the non-legume Casuarina led to the dis-covery of a highly diverged non-symbiotic globin gene in soybean25
. Even when this ancient paralogue is ignored, the gene tree for symbiotic globins of legumes is com-plicated by what appear to be varying lev-els of concerted evolution. This has led to homogenization in some genera, such as soybean and lupin, but cross-species reten-tion of duplicate (orthologous and paralo-gous) relationships in genera such as alfalfa (Medicago) and broadbean (Vicia)22. In the latter genus, the most distantly related member of the gene family was also found to be induced in mycorrhizal symbiosis26 . Given the possible connections between the two symbioses, it will be interesting to see whether orthologues (members of the same subfamily, divergent because of spe-ciation rather than duplication) of this gene can be identified in other legumes, and whether these or other members of the symbiotic clade of leghemoglobin genes in other legume genera are also expressed in mycorrhizal associations.
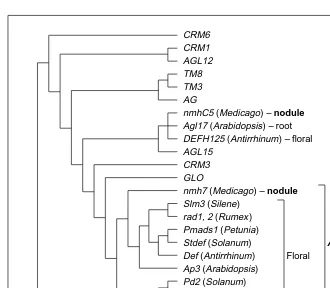
A critical step in understanding the mol-ecular basis of development in any tissue or organ is the identification of specific transcription factors. Among the most in-tensely studied plant transcription factors are the members of the large MADS-box gene family, best known for their role in floral morphogenesis, but also expressed in vegetative tissues of flowering plants and in vari-ous tissues of conifers and even ferns27. Two MADS transcripts have been identified in nodules of alfalfa, and have been found to belong to different MADS-box gene subfamilies28,29(Fig. 3). The phylogenetic relationships of these genes raise more questions than they answer, perhaps illustrating the degree to which recruit-ment to novel functions has occurred in this gene family. There appears to be no common theme among the alfalfa nodule MADS-box gene nmhC5, the root-expressed Agl17 of Arabidopsis to which nmhC5 was found to be most closely related28, and the Antir-rhinum orthologue of Agl17, which is expressed in pollen30
. Simi-larly, the other nodule-expressed alfalfa MADS-box gene, nmh7, is a member of a subfamily that includes Apetala3 of Arabidopsis and Deficiens of Antirrhinum, both B-class floral genes known for their dramatic effect on petal and stamen development. Intriguingly, it appears that no MADS-box gene with detectable sequence simi-larity to nmh7 is expressed in alfalfa flowers29
. Could recruitment of the nmh7 gene product to a nodule function and its lack of expression in flowers be functionally or evolutionarily related? Whether this expression pattern is generally true even of legumes remains to be discovered.
Molecular systematics of bacterial symbionts:
bacterial taxonomy, coevolution and horizontal transfer
Molecular phylogenetic studies are also revolutionizing our under-standing of the bacterial component of the symbiosis. This is not only of great interest to bacterial taxonomists, but is also potentially relevant in elucidating nodule origins. At a coarse scale there is some
Fig. 3. Phylogeny of the MADS-box transcription factor gene family. The basic topology is
from Ref. 27; nmhC5 (Ref. 28) and DEFH125 (Ref. 30) were placed in the AGL17 subfam-ily, but have not been included together in a comprehensive phylogenetic analysis and are shown as unresolved. Genes expressed in Medicago nodules belong to two subfamilies; gen-era from which other genes of these subfamilies were isolated are shown. One of these sub-families (AP3) consists of genes expressed in floral organs, whereas each of the genes of the other subfamily (AGL17) has a different expression pattern.
CRM6 CRM1 AGL12 TM8 TM3 AG
nmhC5 (Medicago) – nodule Agl17 (Arabidopsis) – root DEFH125 (Antirrhinum) – floral AGL15
CRM3 GLO
nmh7 (Medicago) – nodule Slm3 (Silene)
rad1, 2 (Rumex) Pmads1 (Petunia) Stdef (Solanum) Def (Antirrhinum) Ap3 (Arabidopsis) Pd2 (Solanum) Tm6 (Lycopersicon) Cmb2 (Dianthus) SQUA
AGL6 AGL2
AP3 AGL17
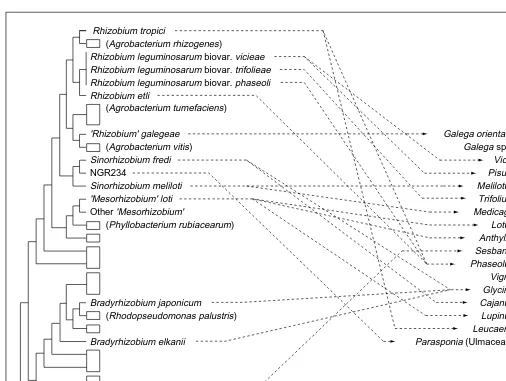
correlation between bacterial and plant relationships, with Gram-negative ‘rhizobia’ participating in symbioses with legumes, and Gram-positive bacteria nodulating most of the non-leguminous nodulators. At a finer scale, if there has been cospeciation of host plants and rhizobia, then the presence of the same or closely related rhizobial taxa in all members of a nodulating group would argue for homology of their nodules; conversely, the presence of unrelated rhizobia in the nodules of different species would argue for inde-pendent origins of nodules. Phylogenies inferred from 16S ribo-somal RNA gene sequences emphasize the great genetic diversity of the bacteria involved in legume symbioses31
. ‘Rhizobia’ include representatives from several major lineages of proteobacteria, with the symbionts in each lineage being more closely related to non-symbionts (such as Agrobacterium tumefaciens, in the case of Rhi-zobium itself) than to symbionts in other lineages31
(Fig. 4). The 16S rRNA gene phylogenies indicate that there is little, if any, phylo-genetic correlation between bacteria and their legume hosts. For ex-ample, Phaseolus, a member of the tribe Phaseoleae, is nodulated by Rhizobium leguminosarum, strains of which also nodulate mem-bers of other tribes. Another member of the Phaseoleae, Glycine, is
not nodulated by Rhizobium, but instead has symbioses with mem-bers of two distinct lineages of Bradyrhizobium and with a Sino-rhizobium species (Fig. 4). The wide host-ranges of some bacteria further complicate matters: a single Rhizobium strain, NGR234, can nodulate not only diverse legumes but also Parasponia.
It is possible that phylogenies based on 16S rRNA sequences do not provide appropriate data for addressing nodulation origins. Bac-terial genes involved in nodulation are commonly found on plas-mids, and horizontal transfer might have played an important role in the evolution of nodulation32. However, not all nodulation genes are borne on plasmids: several key nodulation genes were found to be absent from the fully sequenced 536 kb plasmid of the wide host-range rhizobium strain NGR234 (Ref. 33). However, even nodu-lation genes that are grouped in chromosomal ‘symbiosis islands’ appear to be capable of transmission among diverse bacteria34
. Thus, phylogenetic analyses of nodulation genes can tell very dif-ferent stories from the 16S rRNA genes used to construct bacterial phylogenies32,35. The relevant information on plant–bacterial co-evolution might be gained from these genes or gene complexes, rather than from the bacteria that now bear them36.
Fig. 4. Comparison of legume and bacterial molecular phylogenies. The bacterial phylogeny31
(left) is based on 16S rRNA gene sequences, and shows three very distinct groups of bacteria involved in nitrogen-fixing symbiosis with legumes and Parasponia (Ulmaceae). Representative symbiotic bacteria are shown; lineages of non-symbiotic bacteria are shown by boxes, with a representative given in parentheses for lineages closely related to symbiotic groups. On the right, phylogenetic relationships are shown for selected legume genera (reduced from the tree in Fig. 2) nodulated by bacteria shown on the bacterial tree. Arrows connect bacterial symbionts with their plant hosts. The wide host-range of NGR234 is shown only by indicating its ability to nodulate Parasponia. Although host-ranges of individual bacteria are likely to be wider than shown here, the lack of correlation between the 16S rRNA bacterial phylogeny and the rbcL phylogeny of Leguminosae is still apparent.
Galega orientalis Galega spp. Vicia Pisum Melilotus Trifolium Medicago Lotus Anthyllis Sesbania Phaseolus Vigna Glycine Cajanus Lupinus Leucaena Parasponia (Ulmaceae) Rhizobium tropici
(Agrobacterium rhizogenes)
Rhizobium leguminosarum biovar. vicieae Rhizobium leguminosarum biovar. trifolieae Rhizobium leguminosarum biovar. phaseoli Rhizobium etli
(Agrobacterium tumefaciens)
'Rhizobium' galegeae (Agrobacterium vitis) Sinorhizobium fredi NGR234
Sinorhizobium meliloti 'Mesorhizobium' loti Other 'Mesorhizobium'
(Phyllobacterium rubiacearum)
Bradyrhizobium japonicum (Rhodopseudomonas palustris)
Bradyrhizobium elkanii
Conclusions and prospects
Our understanding of nodulation is indeed evolving, and phylo-genetic studies are playing a significant role in this process. For example, our view of nodulation has changed from one of mul-tiple, completely independent origins in totally unrelated plant groups to perhaps a single origin of a predisposition for nodu-lation. Similarly, nodulins are no longer considered to be uniquely expressed in nodules, but are thought to have been recruited from diverse ancestral functions based on pre-existing genes. Other new insights include an improved understanding of the diversity of relatively distantly related bacterial genera that play a part in nodulation.
Increased phylogenetic sampling of plants will allow hypoth-eses on nodule evolution to be refined. Studies on the expression and phylogenetic patterns of genes involved in nodulation, particu-larly in plants other than the handful already intensively studied, will provide insights into how the nodule was assembled in the course of evolution. In the bacterial symbiont, elucidation of the complex relationships of both bacterial species and particular bac-terial genes affords similar opportunities.
Acknowledgements
I gratefully acknowledge grant support from the US National Science Foundation (DEB-9614984 and DEB-9420215).
References
1 Wagner, G.M. (1997) Azolla: A review of its biology and utilization, Bot. Rev.
63, 1–26
2 Ahern, C.P. and Staff, I.A. (1994) Symbiosis in cycads: The origin and
development of coralloid roots in Macrozamia communis (Cycadaceae), Am.
J. Bot. 81, 1559–1570
3 Gantar, M. et al. (1995) Colonization of wheat (Triticum vulgare L.) by
N2-fixing cyanobacteria: IV. Dark nitrogenase activity and effects of cyanobacteria on natural 15N abundance in the plants, New Phytol. 129, 337–343
4 Swensen, S.M. and Mullin, B.C. (1997) The impact of molecular systematics
on hypotheses for the evolution of root nodule symbioses and implications for expanding symbioses to new host plant genera, Plant Soil 194, 185–192
5 Swensen, S.M. (1996) The evolution of actinorhizal symbioses: evidence for
multiple origins of the symbiotic association, Am. J. Bot. 83, 1503–1521
6 Soltis, D.E. et al. (1995) Chloroplast gene sequence data suggest a single
origin of the predisposition for symbiotic fixation in angiosperms, Proc. Natl.
Acad. Sci. U. S. A. 92, 2647–2651
7 de Faria, S.M. et al. (1989) Occurrence of nodulation in the Leguminosae, New Phytol. 111, 607–619
8 Bryan, J.A., Berlyn, G.P. and Gordon, J.C. (1996) Toward a new concept of
the evolution of symbiotic nitrogen fixation in the Leguminosae, Plant Soil 186, 151–159
9 McKey, D. (1994) Legumes and nitrogen: The evolutionary ecology of a
nitrogen-demanding lifestyle, in Advances in Legume Systematics, Part 5:
Nitrogen Economy (Sprent, J.I. and McKey, D., eds), pp. 211–228, Royal
Botanic Gardens, Kew
10 Doyle, J.J. et al. (1997) A phylogeny of the chloroplast gene rbcL in the
Leguminosae: Taxonomic correlations and insights into the evolution of nodulation, Am. J. Bot. 84, 541–554
11 Kozik, A. et al. (1996) The pea early nodulin gene PsENOD7 maps in the
region of linkage group I containing sym2 and leghaemoglobin, Plant Mol.
Biol. 31, 149–156
12 Ghassemi, F. and Gresshoff, P.M. (1998) The early enod2 and the
leghemoglobin (lbc3) genes segregate independently from other known soybean symbiotic genes, Mol. Plant–Microbe Interact. 11, 6–13
13 Sanderson, M.J. and Wojciechowski, M.F. (1996) Diversification rates in a
temperate legume clade: Are there ‘so many species’ of Astragalus (Fabaceae)? Am. J. Bot. 83, 1488–1502
14 Lavin, M. et al. (1998) Monophyletic subgroups of the tribe Millettieae
(Leguminosae) as revealed by phytochrome nucleotide sequence data, Am. J.
Bot. 85, 412–433
15 Hirsch, A.M. and LaRue, T.A. (1998) Is the legume nodule a modified root or
stem or an organ sui generis? Crit. Rev. Plant Sci. 16, 361–392
16 Gianinazzi-Pearson, V. (1996) Plant cell responses to arbuscular mycorrhizal
fungi: Getting to the roots of the symbiosis, Plant Cell 8, 1871–1883
17 Van Rhijn, P. et al. (1997) Expression of early nodulin genes in alfalfa
mycorrhizae indicates that signal transduction pathways used in forming arbuscular mycorrhizae and Rhizobium-induced nodules may be conserved,
Proc. Natl. Acad. Sci. U. S. A. 94, 5467–5472
18 Franche, C. et al. (1998) Actinorhizal symbioses: Recent advances in plant
molecular and genetic transformation studies, Crit. Rev. Plant Sci. 17, 1–28
19 Verma, D.P.S., Hu, C.A. and Zhang, M. (1992) Root nodule development:
origin, function and regulation of nodulin genes, Physiol. Plant. 85, 253–265
20 Nap, J-P. and Bisseling, N. (1990) The roots of nodulins, Physiol. Plant. 79,
407–414
21 Takane, K.I., Tajima, S. and Kouchi, H. (1997) Two distinct uricase II
(Nodulin 35) genes are differentially expressed in soybean plants, Mol.
Plant–Microbe Interact. 10, 735–741
22 Doyle, J.J. (1994) Phylogeny of the legume family: an approach to
understanding the origins of nodulation, Annu. Rev. Ecol. Syst. 25, 325–349
23 Clegg, M.T., Cummings, M.P. and Durbin, M.L. (1997) The evolution of plant
nuclear genes, Proc. Natl. Acad. Sci. U. S. A. 94, 7791–7798
24 Appleby, C.A., Dennis, E.S. and Peacock, W.J. (1990) A primaeval origin for
plant and animal hemoglobins, Aust. Syst. Bot. 3, 81–90
25 Andersson, C.R. et al. (1996) A new hemoglobin gene from soybean: A role
for hemoglobin in all plants, Proc. Natl. Acad. Sci. U. S. A. 93, 5682–5687
26 Frühling, M. et al. (1997) The Vicia faba leghemoglobin gene VfLb29 is
induced in root nodules and in roots colonized by the arbuscular mycorrhizal fungus Glomus fasciculatum, Mol. Plant–Microbe Interact. 10, 124–131
27 Münster, T. et al. (1997) Floral homeotic genes were recruited from
homologous MADS-box genes preexisting in the common ancestor of ferns and seed plants, Proc. Natl. Acad. Sci. U. S. A. 94, 2415–2420
28 Heard, J., Caspi, M. and Dunn, K. (1997) Evolutionary diversity of
symbiotically induced nodule MADS box genes: Characterization of nmhC5, a member of a novel subfamily, Mol. Plant–Microbe Interact. 10, 665–676
29 Heard, J. and Dunn, K. (1995) Symbiotic induction of a MADS-box gene
during development of alfalfa root nodules, Proc. Natl. Acad. Sci. U. S. A. 92, 5273–5277
30 Zachgo, S., Saedler, H. and Schwarz-Sommer, Z. (1997) Pollen-specific
expression of DEFH125, a MADS-box transcription factor in Antirrhinum with unusual features, Plant J. 11, 1043–1050
31 Young, J.P.W. (1996) Phylogeny and taxonomy of rhizobia, Plant Soil 186,
45–52
32 Mergaert, P., Van Montagu, M. and Holsters, M. (1997) Molecular
mechanisms of Nod factor diversity, Mol. Microbiol. 25, 811–817
33 Freiberg, C. et al. (1997) Molecular basis of symbiosis between Rhizobium
and legumes, Nature 387, 394–401
34 Sullivan, J.T. and Ronson, C.W. (1998) Evolution of rhizobia by acquisition
of a 500-kb symbiosis island that integrates into a phe-tRNA gene, Proc. Natl.
Acad. Sci. U. S. A. 95, 5145–5149
35 Haukka, K., Lindstrom, K. and Young, J.P.W. (1998) Three phylogenetic
groups of nodA and nifH genes in Sinorhizobium and Mesorhizobium isolates from leguminous trees growing in Africa and Latin America, Appl. Environ.
Microbiol. 64, 419–426
36 Ueda, T., Suga, Y., Yahiro, N. and Matsuguchi, T. (1995) Phylogeny of sym
plasmids of rhizobia by PCR-based sequencing of a nodC segment,
J. Bacteriol. 177, 468–472