I
ET
I
I
I
I
I
I
33
SClmago
Journal & Country Rank
EST MODUS IN HEBUS ierii,s isatl( I - 1.1061
; -'nl9
Journal
SearchScarchqury
in jc!:,_1di T1li,. - :en.*
Crunby RanHnBs
Country Search
Conpare
,,lap &nerator
Help
About Us
lnOraitnr lU|r --!r14
sJR aa '
a a a
cit6 t a
perdoc""'
foufiirv; : Sub;ect Afedr
5ubje.t Categcrvi
Biochemistry, Geft etics aft d Molecular Biotogy
{miscellaneoE)
PrSlists: Elsevier Bv. Pt&li.nfist r}?er -hr8els. ISSN: 2221t691
fawqe:2fiI-201,t
H lndex 23
Quartit! iQ1 ffr:fi! highest,.sluEs :,1d q+ l36est ,.eluesl
i r t.drr
19..! :Lrlri :!tt :.(a.l .rf l :ia.t :li-l .ir,-ri :r!l i,inf :i!: :,11t1 ;utt iuli :Llll :Llt.t
0.59
1,68
EES
, r : 16 \ l!
Show thit ,nformation in your own website
Jtr;-u:rai \Ietr-ics
,'
l: - ' ;
SCl mego Jourita I Ra nk (SJR! o.5ty
o
Stayup-t+.date
R€gister ylur inter€Ets rnd r€c€iye email alerts tailored toyour neetls Click here to cignup
Asian
Pacifi.c
Journai
of
Tropical
Biomedicine Editor{al
Board
Editorin
Chief
I€ffieyIvLBetlony
Georp Washington lhintrsity, USA SantiagoMas-Coma
Uhirrrmidrd
1e
ValenciL $p{in
Itfialcolm
It
lones
Queensland Irstihrte of Medical Rrsear,&,,Austualia Vanessa$fgenk
rnp
thiversity of hetori+ South Afiica
Nauni Bte
I{j-
WasiAhmad
Inftctious Disea* Rseanch C,,enteq,
lfiahFir
President Executive
Editor*in-(hief
ShmldaiQu
llainan Medical fhi?Ersity, Chire
|ong-YilChai
Seoul National thircrsity College of Medicing Komn
StepheuMunga
I(mye Medicel Rescarch Institute,
IftDp
ChristopherV- Plowc
(hiwrsity of lrlardend Schml of Medicin€, {rSA
Pierrr Roques
IrEtitute for F.merghg Diseeses ard lororetine Theepieq Frmce
ffi
cuia.forAuthors
v
trlh
submitvour PaperAsian
Pacific
Journal
of
Tropical
Biomedicine
p-ISSN:
[Asian
Pac
J
Trop
Biomedl
2221-r6e1
ICV
2014:122.47
Standaxdized Value: 7.66
at"u-
lVOg*l-gEtegg"t
Previous evaluation
IC
Print version: yesElectronic version: no
I}-:-,r-""-*r*:l
H ir"9-qr":9-i*
i
Biological
Abstracts,
BIOSIS PreviewlBIOSIS,
Cambridge Scientific Abstracts (CSA,
Proquest),Chemical Abstracts (CAS),
EMBASE, IC
JournalsMaster
List,
PubMed/PubMed
Central/lvledline,
Scopus,Ulrich's
periodicals, Zoalogical
RecordBasic
information
Main
title
[English]
*::t
Pacific
Journal
of
rropical
Biomedicine
English
title
*:lTii:,*
Joumal
of
rropical
tJlomeotclneAbbreviated
title
Asian Pac J Trop Biomed
Previous
title
No
p-ISSN
2221-1691
URL
|t-tp;ji*tr,i:s:.-x,i
DOI
10.1016Country
ilChina
(mainland)
Hainan Medical
University,
Asian
Affiliation
Pacific Tropical Medicine
Press,Haikou,
HainanAffiliation country
flChina (mainland)
Journal'scharacter
scientific
Frequency
Monthly
Circulation
3000Asian Paciflc Journai of Trcprcai Biomedicine
€ FeYioG mlJis Next rolfi$ > ,9.'t.;r I - 17
Artides in Press
Ope[ Aeess ailicles
Vdum6t-6{2oIt-2016)
Vdume 6, lssE4
= pp. Z7S-370 iAp{il 2016) *
Volume 6, l$E 3 E pp. 18+2?8 iilaffh ?016]
VolumeS,Ewz E
pF. S3-184 Se'.xq- 201€) Volume 6,1sre 'l
= pp. 1 -92 (JanuaB' 201 E) Volume 5. lsue '12
= pp. 977-10E4iDEemaer2015) * Volres, Eue ll
= pp. 88+!76 iN.*mbBr 2B'15) Volumes lsElo H pp. 789-88.t (O€iober m15)
Volume 5, lse I
= pp. E5+788 (SeptEmber2015! Volrrmps lslcB
= pp- 601-6b4 (Auqust2015) t Volumes lsueT
=
Fr 50$600 (Juif 2ol5)
-Volumes lsw6 E
pp. 421-508 [June 2015j, * Vdumesl$ues tr pp.337-420 th{ar 2015}
Volume 5, lsue4
= pp. 253-336 P,pril 201 5) Volumes rsreS
= pp. 169!52 {t.a{dr 2015}
Volume 5, lsuez
= pp. 8''168 (Fehrusrj 20151 Vofure6,$uei
= pF. 1-84 (Jiluae" 2015)
Yolume4,l$ue 12 = pF. 935- 1008 (O"cember 201 4]
Volume4,lse 11
= FE. 941-92,1{Ntr+miH 20141 "
Volume4,lssG'lo E pp- 757-840 {Odober 201 4) Volume 4. l$E g
= pp E7:-7id (SBdember2014)
-Volume4, l$EB
= pp. 5A$d72 {tugust 281 4)
Volume4, lsueT E pp- 58+588 (July 2814i
Volume4 lsue 6 tr
!B 4?1-50{ iJune 2010, *
Votume4 lsueS E Fp. 337-420 (hlay 2814)
Volume 4 lsue4
= pp. 25+336 (.Aprjl m1+) -Volume4 lsre3
=
,r. 1E!-25? iLisch 2014)
Volured tsre2
= pp.8F168 {Februaryml4) Volume4 lsE'l
=.j pp. 1-81(Jmary 2014) Volume4 Supplffient2
= pp S53s-s682 (Julji 2B t4)
t
f opoa*"-tro'
]E Crypto6IoririGb a|lMg driHren with dierhGa in three r6ian Mtries: A Hiewtuka.* fu@WN
B.*E Lfif, !&ffrFeiza R*
r llsa'a G PDrt44gE
tr ConfrrctuUclAaogizlgEntuatu!teaertungidqfrse,*d*riimHeifiEFed OpsnffiE
iEHIy f*edih{rffi h.&b€ ter6at6na6aG* em sodrexn Hy oiqid Fbffidr Ar{ide
PerBe,9.a4S
ffiB tanqtE lkia P,ia Pasr&i. Wa Xiz4 l(atE lraafirdq t*auilto FmirE $HrBr Co6ts0@,
Gtrrdo Fabre, Fh.ffiFsfo"eBldo k*e6q, LrisOe lila&E
l ADEr^*r E poEtsmrq
fl Clfiorqijtyev*etimof exrractsaldftactimoffireffinespoagEfrwtEI%E!ilGlf sd OegrlrcE HPLC fingsprint s$atFiE of cltotub extracb cilin Rffidr arHe
@N@r
tlaEdl&hfiflf t$Hlr#y,rffir$el,ca.Hos€iri, tt&Yffii, Ue6altaEtri, tlE.sda.tftEfl**
r emxr S PDF(11szre
E lrlobdhrstudyan r€thkiltrrc&tsnt fugr@E il,ffisfraiE imlat€d frutr dogsaftd opsre* E
sociated personnel ia Jordan o{ignrd frEffidr r.*
fuEw-*,
Yffi fla:l#rIffii,rfiBEdl8rrEdISffiFl&l*G&*olmfilad,(lH,cbabaear,taffi SlErreulx'red,
A.fH SdHn Jar{ r *ra-a* G P!FE53$
E ER6TrmutatircmiateduilhaabcEErein fund;&afu*m'EE,latsfrmwhovqiEt qslffiEl €rldircb patients orbid RMdl rrEE
P#gl94r1
tEnW*E,Li+*Ef*sE.FXFZm,fa*rfY€(l*stilro,DrFYaEItlEEt**EFfattraEt.n€ilEYnl|d](slg
)Absfd.:fi
eortusnE EEtrEfinof z@rEticpotsEydEsdrai*Eef0lszFrl fuq€fiarbitrantyprinredPCR o*siffi= ne*hodsOrEid fknfi add.
@egl,W
I tithfa.t gBdaE, f)!E,rfyuWI(HI l{}I*ihE*rKr*Ueffi, l(ilECJtlsr}aFlrirafk*\*dlselaf
EtryHE
r Looi"d E PDFFffitg
E Art*rnarlretoryryd entipyrelicpropertiBof Kffig 601 h€ji, a lraditiffil Chiworal tquid tuge opsffi= fomorifdRmdrffie
fu@Af-*z
HEdHanm*ere*mea, 5T$Wgvottl*Bf,UAE&riitAslDtsGatft Affidulfb6ia4EDe S
trctf#am
r .Absr*t fr PDF(gSl rg
E tueeEnerielactivityof fiv€Pewiilm€diiHlplsnl3qainst Mtunawsztgi@ortidtusrrr o!.nAffiEl
*fiG
ru@#E-*1,
csbieaatxtm{.xi?tr,IdorrdliudAguB{-Lr*x lff3.tdC*ffiDeLrE{ilAJoe€crrffi-LErdrq
.IEE del\iBE ItsS@ r rouara G P9Ft311i0
E tuti#la{ial €tred of I -(N#tyLaffihtryl)-stdrqF2f,elhtEyridin-rt-ore and glem ts op* r* El ecdr*l tr erythrccyHage Pbffiim fugha' in mhe orisid faffi{fi Frnce
@@et
Frr*$EThip.So.L rtd*rrymIiiffi, ClEir*tltEBdx$, gtEstSilEielal, Sqndetgijffitffi
r *araa E PDF(.r74tQ
E Wffrd hedng potffitial ot rryhe ofrirefsfforermrikge inr*bitfult thiiosrcundsffqid openAffi=
REddr aasch w#7-91i1
RobbVafafttlrE HiElMi,GtEbM;rqM,SaaBald,ltM Bdffi*
Volume4, Supplem&t 1 = pp. Sl-S53S ilrlay 2014]
Volume 3, lse 1? = p!. S25-1012{DeEm}er2013} "'-Volume g, Tsue 11 .:i !F. S41-934{NovemSer ?01-?l
-'-Votume 3, $@ 10 i pp. 7514,t0 iodnSer 4013i
Volume 3, sue 9 I.l
pp. 6??-795 (Septemier 20131
Volume 3, lsseS :Z
pF. 589-572 {.4ucust 3013}
Volume 3, l*ue 7 '.:L !p. 50SSS$ Uuly 2013) volume 3. lsue 6 l':r' pp.4?1-504 iJune 2t13)
Vdume 3, lsue 5 i::r BE. 337-420 {l,,la! 2013} Volume 3, lse4 := Fp. !53-336 P.lril ti I 3) Volume 3, lsre 3
-pp. 169-252 (i,tardi !013i
Volume 3, lsw 2 H F!. B+168 {Fetruary 2!13}
Volumes.lsre 1
= Bs. 1-g4ljauarl1013)
Volume!. lssue 1l
FF Ia7-8i'l iC':ti:er !81::
Volume 2 l$ue g FF El:-7aE isgFteri:*r !!11:
'{slume 2. lssue.q pF. arlg-Li7: {ru,!ust irll:i
Vslume, lssue 7 PF ai:-a8i LJult !i]lIi \isluflE ! lssle 6 FF r:'l-arlj lJunE 2012: Vdume 2, lssue 1? .:::. Fp !2i-10t8 iDecemler !['!lj Volume 2. l$ue '1'l :,::: FF. B.i1-g!4if',lrvember2012i''
m Cyotffie adirrityard pttlltoctEfnicd an#is of nrmpeAedimr8otss. orEirdRffidredde
ru@!w..ui
ila.i ilolmryred Feid, Sffid) Re{k HEsdn, Lffiiffi FaE/ lbr**m, t*ltrffimed EHrEd,afifiEdl&El Od*l, tlalxrBrdleaEH€riSds
r eosrad G PDFt506rq
El Dcr*ing c-syndein a€grsgetion by retEmlic erdtrt of delrte& ffibl*r in zebEfEtl Perldl1s's
flEdel OrhEd FlEwdl &liiie
w9r8w
lhmd HlEltwh, UuBEhadi Ati.g.*wrBa*lailq s,mlko, Ldffiad AisW rlo(b ) AD6&act E por(rmrrq
E A rwd HPTLC nrelhod ior qrentitatire estimtim of *narlqs in pdyterbel fomulation o'tid ReffidrtHc
fiagesC$!rB
ZeslEntam€dSrEikh, SadaShslcd, SmnEqr,l{ibza,ffi, Sak*EStrsrBrcEl, FaisdlffiZ;*di, t$H f.jsH$mi
)i6sFad E PDF(60riq
fl Chsnisl mmpeitim and etihactsial acti\rity ol ffitiel dl of I aw &,nifsa Pau gIffi in F.lgedan ari, step* odsid nffit*t rrtbb
fues@-S4
Tsei( B€mEdotr, l+ocirE Lffi, Saffir Adcd, &rdo Fl#*ni r,rmrract fi PDFcSrcQ
El flkfi Burgm of dengre 1Ector popuhtixe *ter spee spEyirts in il sdemb urhan ro of
Thailand: A cfusler Endomized mtafied trialo'ilid rbEdt rrr&h ruwffi-gm
N4a{rd S!Mn,KrJBffiTtrt}aito, Slsid}]}mfl@b,VirffiMiCtEng$fliutEE,nMabrdPsrgsisj
) AbstEc.r E PDF(100$$
E Odendmle a sffisirl treatrenl Dt TaHb hldatbwa reaastod* in mtuBlty infect€d piF fu6sn-973
Luis Anbnio Go(@+rsb, Ameldo BniIim Gonzde, Csrcfl*da, \f#bo l#w, H€#itugo Gtrcia, Mffia Tffi Lop€a-UrliE, q/drmtsn*irg GrqpinPffi
) Aestrtrr & PDF(.r16lQ
E Matlrgratical mode] erytainirlg ircn change iE patiHts uith thdmia
fut$97+975
Ssn$i lvi$B*ril Vircj i,Viw*tHt
fi
eor(m+lgoetrt*cffiEl
oF@lffi=
tulid* t - 17
Original article http://dx.doi.org/10.1016/j.apjtb.2015.07.023
Evaluation of zoonotic potency of
Escherichia coli
O157:H7 through arbitrarily primed
PCR methods
I Wayan Suardana1*, Dyah Ayu Widiasih2, I Gusti Ngurah Kade Mahardika3, Komang Januartha Putra Pinatih4, Budi Setiadi Daryono5
1Department of Veterinary Public Health, Faculty of Veterinary Medicine, Udayana University, Jl. PB. Sudirman Denpasar,
Bali, Indonesia
2Department of Veterinary Public Health, Faculty of Veterinary Medicine, Gadjah Mada University, Jl.Fauna 2, Karang
Malang, Yogyakarta 55281, Indonesia
3Department of Virology, Faculty of Veterinary Medicine, Udayana University, Jl. PB. Sudirman Denpasar, Bali, Indonesia 4Department of Clinical Microbiology, Faculty of Medicine, Udayana University, Jl. PB. Sudirman Denpasar, Bali, Indonesia 5Department of Genetic, Faculty of Biology, Gadjah Mada University, Jl. Teknika Selatan, Sekip Utara, Yogyakarta 55281,
Indonesia
A R T I C L E I N F O
Article history: Received 25 May 2015
Received in revised form 24 Jun, 2nd revised form 20 Jul 2015
Accepted 26 Jul 2015 Available online 30 Aug 2015
Keywords:
E. coliO157:H7
Zoonoses Animals Human AP-PCR
A B S T R A C T
Objective: To evaluate the zoonotic potency of Escherichia coli O157:H7 through arbitrarily primed-PCR (AP-PCR) methods as one of the DNAfingerprinting methods. Methods: A total of 14 isolates consisted of 11 isolates originated from human feces with renal failure symptoms, 2 isolates originated from cattle feces, and 1 control isolate were used in this study. DNA of each isolate was extracted, and their profiles were studied by using AP-PCR method with M13 F and M13 R arbitrary primers.
Results: The results founded that all of 14 isolates had similarity range from 54.6% to 88.5%. Isolates KL-106(3) and KL-55(6) originated from humans showed the degree of similarity with isolates SM-25(1) and SM-7(1) originated from cattle as high as 85% and 77%, respectively.
Conclusions: The high degree of similarity between isolates originated from cattle and human indicated the high potency of zoonoses. The results also concluded AP-PCR method as a briefly fingerprinting method in order to trace the epidemiological of
E. coliO157:H7.
1. Introduction
Escherichia coli(E. coli) O157:H7 is a zoonotic agent of the
type of Shiga toxin-producingE. colithat can cause disease in humans at some parts of the world[1–4], and cattle are known as the
main reservoir of these bacteria[5,6]. Researchers had succeeded in isolating these bacteria from pigs, beef, pork, water or human[7,8]
and proved the food products obtained from supermarkets were contaminated withE. coliO157:H7[9,10].
The infection by these bacteria in animals is usually asymptomatic, whereas these bacterial infections in humans usually show clinical symptoms of diarrhea and hemolytic ure-mic syndrome [1,11,12]. The human infection usually occurred
when proper hygiene is not strictly implemented especially in many developing countries and human consumed undercooked food products[2,13].
In Indonesia, several studies related toE. coliO157:H7, have been carried out by researchers. The study of Drastini in 2007 had found that 59% of vero toxinE. coliconsisting of vero toxin
E. coli O157 and non-O157 can be isolated from all types of
livestock namely, dairy cattle, beef, pigs and goats/sheep [14]. Other researchers reported theirfindings ofE. coliO157:H7 in Badung, Bali in beef and in healthy human feces were 5.62% and 1.30%, respectively [15]. The slight titer toxins of Stx1
and Stx2 resulted from local isolates ofE. coliO157:H7 were
*Corresponding author: I Wayan Suardana, Department of Veterinary Public Health, Faculty of Veterinary Medicine, Udayana University, Jl. PB. Sudirman Denpasar, Bali, Indonesia.
E-mails:[email protected],[email protected]
Peer review under responsibility of Hainan Medical University.
Foundation Project: Supported by the Directorate of Research and Community Services, Directorate General of Higher Education through Udayana Research Grants with contract No. 21.34/UN14/SBRC/2012.
H O S T E D B Y
Contents lists available atScienceDirect
Asian Paci
fi
c Journal of Tropical Biomedicine
journal homepage:www.elsevier.com/locate/apjtb
Asian Pac J Trop Biomed2015; 5(11): 915–920 915
originated from cattle feces and beef had been reported [16]. Moreover, the pheno-genotypic adherence ofE. coli O157:H7 isolated from beef, feces of cattle, chicken and human had also been studied[17,18].
According to theory, the detection of pathogenic microor-ganisms including E. coli O157:H7 was preferable to using molecular methods, considering the highly sensitivity and specificity of these methods. Molecular method in order to study the epidemiology of food-borne diseases is usually using DNA subtyping. Genomic DNA profiles in DNA subtyping are divided according to the size, and the resulting size will form the patterns which can be used as a characterization of each isolate
[19]. Some DNA subtyping methods are commonly used for the
characterization of bacterial genomes including pulsed-field gel electrophoresis, ribotyping, and random amplified polymorphic DNA (RAPD)[20,21].
Pulsed-field gel electrophoresis subtyping as one of the
fingerprinting methods usually use restriction enzymes that cut genomic DNA randomly to generate approximately 20–30
fragments. The size of the fragments formed quite varied be-tween 10 and 1 000 kbp and this method requires a separation method with special electrophoresis[19,20]. On the other hand, ribotyping method also uses restriction enzymes. Restriction enzyme will cut the DNA fragment in the gene encoding ribosomal RNA. The number of fragments produced will vary ranges from 500 to 1 000 fragments with measure between 1 and 20 kbp. This method has limitation because the results can only be detected using a specific probe for the gene encoding ribosomal RNA from bacteria[22].
RAPD method was developed by researcher using short ol-igonucleotides (10 bases) as random primers. This method was successfully performed by researchers to differentiate the bac-terial genome with only a small amount of genomic DNA[23,24]. RAPD method had been successfully used in an effort to study the zoonotic potency ofE. coliO157:H7[25,26].
In the development of this method, the other researchers developed a similar method that used approximately 15 nucle-otides as random primers with different amplification conditions with RAPD which is known as the method of arbitrarily primed PCR (AP-PCR). The use of AP-PCR method as a variant of RAPD method for epidemiological investigation has been widely used by researchers[27,28].
AP-PCR method is known to have some advantagesi.e. it can be applied to a wide range of organisms, fast and simple to generate fingerprint products. This method also used the selected primers without needing the initial information about the organism to be tested. Besides those, the data generated would be able to provide information about the difference or similarity of strains originated from the same species [29,30]. This method has been success to identify the number and the distribution of genotypes of Streptococcus mutans and
Streptococcus sobrinus [31], to predict the degree of
malignant cholangiocarcinoma as familiarly as the malignant neoplasm of biliary epithelium [32], and to describe an outbreak caused by extended-spectrum beta-lactamase pro-ducing Klebsiella pneumoniae [33]. Furthermore, this method
also proved in good agreement with the results obtained for the 16S rRNA[34].
Based on these facts, in this study we reported the application of AP-PCR method in order to study the zoonotic potency of
E. coli O157:H7 from animals to human and all at once as a
clarification of previous study, which analyzed the zoonotic
aspect of E. coliO157:H7 both animals and human origin by analysis of protein profile and 16S rRNA gene[35,36].
2. Materials and methods
2.1. Bacterial strains
Bacterial strains ofE. coliO157:H7 that used in this study were the same as those used previously[25,36]. Fourteen isolates
consisted of 11 isolates originated from human feces with renal failure symptomsi.e.KL-52(7), KL-87(7), KL-30(4), KL-45(1), 48(2), 85(1), 83(5), 24(5), 68(1), KL-106(3) and KL-55(6), 2 isolates originated from cattle fecesi.e. SM-25(1) and SM-7(1), and 1 control isolatei.e.ATCC 43894 were used in this study.
2.2. Cultivation of bacterial strains
Cultivation of the bacterial isolates was done identically with previous study [35]. Fourteen isolates ofE. coliO157:H7 were taken from stock (stored in 30% glycerol with a storage temperature of −20 C) to subsequent cultivate on lactose broth medium at 37 C for overnight. Isolates were
re-confirmed by culturing on selective medium sorbitol MacCon-key agar, followed by testing on latex agglutination test O157 and testing on H7 antiserum as afinal confirmation.
2.3. Extraction of DNA and PCR
Bacterial DNA was extracted using QIAamp DNA mini kits (Qiagen) according to supplier's instruction as described previ-ously [25,36]. The concentration of DNA was determined by
spectrophotometer. Amplification of genomic DNA by using AP-PCR method was carried out in 40
m
L reaction volumes.These reactions contained 2
m
L DNA template (200 ng/m
L),36
m
L PCR Supermix 2× (Invitrogen), and 2m
L (20 pmol/m
L) of each primer. The primers used were M13F: 50-TGTAAAAC-GACGGCCAG-30and M13R: 50
-CAGGAAACAGCTATGAC-30[37]. PCR program was performed on Eppendorf mastercycler personal. The PCR amplification had initial DNA denaturation at 94C for 5 min, followed by 39 cycles of denaturation at 94C
for 1 min, annealing at 35C for 1 min, and extension at 72C
for 1 min;finished by afinal extension at 72C for 5 min at the
end of the amplification. About 5
m
L of PCR product was analyzed by electrophoresis (Bio-Rad) in 1.5% agarose (Gibco BRL) gel, at 90 v for 45 min. The gel then stained by 1% solution of ethidium bromide (50m
L/L) and destained with tetrabromoethane 1× for 20 min. Gel was visualized by UV transillumination and it was recorded by digital camera FE-270 7.1 megapixels.2.4. Data analysis
The amplified fragments were scored in a descending manner from higher to lower molecular weight in a binary code. The appearance of product was designed as 1 and its absence as 0[38].
Evolutionary distance was measured with algorithm of
unweighted pair group method using arithmetic averages (UPGMA) and similarity coefficients of each cluster were showed near the branch of phenogram. The data were processed using multivariate statistical package 3.1 program[35,39]. I Wayan Suardana et al./Asian Pac J Trop Biomed 2015; 5(11): 915–920
3. Results
3.1. AP-PCR profile
The purified DNA of 14 isolates of E. coli O157:H7 including control isolate was subjected AP-PCR and different patterns of genomic DNA were obtained. The DNA profiles of 14 isolates generated using primer M13F were shown in
Figure 1.
The use of primer M13R showed different patterns. The complete AP-PCR profiles of 14 isolates of E. coli O157:H7 including control isolate ATCC 43894 using primer M13R were shown inFigure 2.
According toFigures 1 and 2, the bands that were obtained from 14 isolates spread out the size range from 300 to 2 000 bp. Total bands and fragment variations of the 14 isolates ofE. coli O157:H7 generated by AP-PCR using both primers M13F and M13R were summarized inTable 1.
3.2. Phenogram analysis
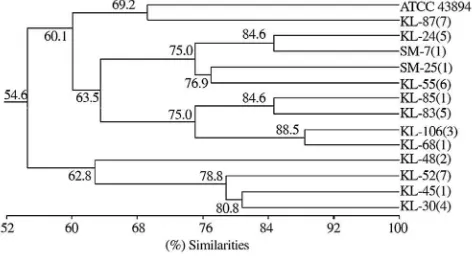
Total bands and fragments of all isolates obtained using AP-PCR method in Table 1 showed variation among isolates. These variations were specific for each isolate so that they can be used to differentiate between one isolate to others. Further analysis of the data in Table 1showed the genetic relatedness among isolates. Genetic relatedness described as a tree-like structure was known as phenogram presented inFigure 3.
Figure 3 shows that E. coliO157:H7 strains were divided into some clades with each coefficient similarities. Isolate KL-87(7) showed closely related to ATCC 43894 with a
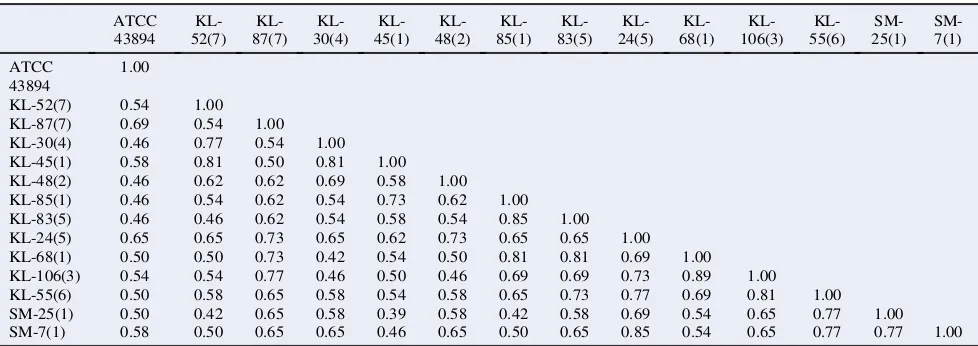
coef-ficient similarity of 69.2 or having similarity of 69.2%. More-over, both KL-45(1) and KL-30(4) isolates shared clade with coefficient similarity of 80.8 or having similarity of 80.8%, as well as the others. The complete matrix of similarity coefficient among isolates ofE. coliO157:H7 was presented inTable 2.
Figure 1. AP-PCR profile of genomic DNA ofE. coliO157:H7 by using primer M13F on 1.5% agarose gel.
Line 1: ATCC 43894 (positive control); Line 2: KL52(7); Line 3: KL87(7); Line 4: KL30(4); Line 5: KL45(1); Line 6: KL(48(2); Line 7: KL85(1); Line 8: KL83(5); Line 9: KL24(5); Line 10: KL68(1); Line 11: KL-106(3); Line 12: KL-55(6); Line 13: SM-25(1); Line 14: SM-7(1); M: Marker 100 bp DNA ladder.
Figure 2. AP-PCR profile of genomic DNA ofE. coliO157:H7 by using primer M13R on 1.5% agarose gel.
Line 1: ATCC 43894 (positive control); Line 2: KL52(7); Line 3: KL87(7); Line 4: KL30(4); Line 5: KL45(1); Line 6: KL(48(2); Line 7: KL85(1); Line 8: KL83(5); Line 9: KL24(5); Line 10: KL68(1); Line 11: KL-106(3); Line 12: KL-55(6); Line 13: SM-25(1); Line 14: SM-7(1); M: Marker 100 bp DNA ladder.
Table 1
Fragment and total band of 14 isolates ofE. coliO157:H7 generated by AP-PCR using primers M13F and M13R.
Isolates Source Fragments with primer M13F (bp) Fragments with primer M13R (bp) Total bands ATCC 43894 Human feces control 300; 450; 550; 600; 700; 2 000 750 7 KL-52(7) Human feces 300; 400; 450; 550; 750; 1 400 400; 450; 550; 800; 850 11
KL-87(7) Human feces 300; 400; 500; 700; 800 – 5
KL-30(4) Human feces 300; 400; 450; 750 400; 450; 500; 550; 700; 850; 1 200 11 KL-45(1) Human feces 300; 400; 450; 600; 750; 1 400; 2 000 400; 450; 500; 550; 850 12 KL-48(2) Human feces 300; 500 400; 450; 500; 550; 700; 800; 1 000 9 KL-85(1) Human feces 400; 500; 600; 800; 1 400; 2 000 400; 450; 500; 550; 900 11 KL-83(5) Human feces 400; 500; 600; 800; 1 400; 2 000 400; 550; 700; 900; 1 200 11
KL-24(5) Human feces – 400; 550 2
KL-68(1) Human feces 400; 500; 600; 800; 1 400 350; 550; 900 8 KL-106(3) Human feces 400; 700; 800; 1 400 350; 550; 900 7 KL-55(6) Human feces 400; 700; 900; 1 400 400; 550; 700; 900 8 SM-25(1) Cattle feces 400; 700; 900 550; 600; 700; 1 000; 1 200 8 SM-7(1) Cattle feces 700 300; 400; 550; 700; 1 200 6
Total general 116
Figure 3. Phenogram ofE. coliO157:H7.
Phenogram was constructed using simple matching coefficient (Ssm) and algorithm UPGMA based on 116 AP-PCR fragments generated by arbitrary primer M13F and M13R.
4. Discussion
AP-PCR as one of thefingerprinting techniques was known to have many advantages such as its simplicity and shorter time consumption. This technique allows genetically different bac-terial strains to be distinguished with great sensitivity and effi -ciency [28]. Sensitive and efficient technique for typing
pathogenic microbes was important for tracing routes of infection and for understanding the spread and evolution of virulence agent [28,34]. In our study, AP-PCR was used as a technique to study zoonotic potency of E. coli O157:H7 in humans from animals' origin.
Our study showed AP-PCR technique as a powerful discrimination method to distinguish each isolate characterized by many bands or fragments that were amplified. Either primer M13F or M13R produced a number of lengthy fragments (Table 1). Total amplification produced by this methodi.e.116 bands consisting of 59 amplicons for primer M13F and 57 amplicons for primer M13R. The lengthy fragment resulted from both primers also showed variation. As many as 12 fragments were detected from primer M13F consist of 300, 400, 450, 500, 550, 600, 700, 750, 800, 900, 1 400 and 2 000 bp. Moreover, primer M13R detected 13 variations consisting of 300, 350, 400, 450, 500, 550, 700, 750, 800, 850, 900, 1 000 and 1 200 bp. The variation in the length and number of fragments was amplified from both primers for each isolate indicated the highly power discrimination of this method as one of the DNAfingerprinting methods to describe the genetic variation for each isolate.
Glick and Pasternak revealed that the primer or short oligo-nucleotides used in genomic analysis would form pairs in many places with chromosomal DNA. The number of amplified DNA fragments would depend on the primers used, and the result could be used to determine the characteristic of the entire genome or the chromosome of an individual[19]. Accuracy of
AP-PCR as one of the fingerprinting methods with a great sensitivity and efficiency had proved by researcher[40]. Their research reported this method was successfully used to distinguishE. coli O157:H7 strain using template from boiled stationary-phase cultures, without the need for time-consuming phenol extraction[41].
The use of AP-PCR method also successfully investigated clonal relatedness among the strains of epidemic isolates of
Vibrio cholerae O1 recovered from an outbreak occurring in
different parts of Iran [42] and to type Clostridium difficile
isolated from different sources of Imam Reza hospital, Tabriz, Iran in order to control nosocomial infections produced by these bacteria [28].
Further analysis of the data in Table 1 showed the genetic relatedness among isolates of E. coli O157:H7. Genetic relat-edness were described as a phenogram in Figure 3showed 14 isolates tested exhibiting clustering into 13 clades. Clade 1 was formed by isolates KL-106(3) and KL-68(1) with 88.5% simi-larity; clade 2 was formed by isolates KL-85(1) and KL-83(5) with 84.6% similarity; as well as clade 3 was formed by iso-lates KL-24(5) and SM-7(1) with 84.6% similarity; clade 4 which was formed by isolates KL-45(1) and KL-30(4) was known to have similarity value of 80.8%; while clade 5 was formed as a joint of clade 4 with isolates KL-52(7) having 78.8% similarity and so on to clade 13 with a value of similarity 54.6%.
Phenogram on Figure 3, which was clarified by similarity values of each isolate that was showed inTable 2exhibiting 2 out of 13 local isolates ofE. coliO157:H7i.e.isolates KL-24(5) and KL-87(7), respectively were known to have lower similarity i.e. 65% and 69%, respectively against control isolates ATCC 43894, which was known as the cause of humans' outbreaks in Japan. On the other hand, some isolates of human were also known to have a high similarity against isolates of cattle. Isolate SM-7(1) showed high similarity against isolates of human i.e. KL-24(5) and KL-55(6) as high as 85% and 77%, respectively. Isolate SM-25(1) also showed high similarity against isolate KL-55(6) originated from human with the degree of similarity 77%. The high degree of similarity between isolates of human and cattle origin, especially for isolates with degree of similarity 70% or more, it could be categorized as the same strains. This view was supported by Rosello-Mora and Amann who revealed in generally, organisms in prokaryotic species could be grouped into the same strain if they have similar genome as high as 70% or greater[43].
The high similarity of genomic DNA between isolates of
E. coliO157:H7 in human and cattle origin indicated its potency
as a zoonotic agent which transmitted from animals especially cattle as a main reservoir to humans. Researchers stated that cattle as a natural reservoir of E. coli O157:H7 could be
Table 2
Similarity coefficient among isolates ofE. coliO157:H7. ATCC 43894 KL-52(7) KL-87(7) KL-30(4) KL-45(1) KL-48(2) KL-85(1) KL-83(5) KL-24(5) KL-68(1) KL-106(3) KL-55(6) SM-25(1) SM-7(1) ATCC 43894 1.00
KL-52(7) 0.54 1.00 KL-87(7) 0.69 0.54 1.00 KL-30(4) 0.46 0.77 0.54 1.00 KL-45(1) 0.58 0.81 0.50 0.81 1.00 KL-48(2) 0.46 0.62 0.62 0.69 0.58 1.00 KL-85(1) 0.46 0.54 0.62 0.54 0.73 0.62 1.00 KL-83(5) 0.46 0.46 0.62 0.54 0.58 0.54 0.85 1.00 KL-24(5) 0.65 0.65 0.73 0.65 0.62 0.73 0.65 0.65 1.00 KL-68(1) 0.50 0.50 0.73 0.42 0.54 0.50 0.81 0.81 0.69 1.00 KL-106(3) 0.54 0.54 0.77 0.46 0.50 0.46 0.69 0.69 0.73 0.89 1.00 KL-55(6) 0.50 0.58 0.65 0.58 0.54 0.58 0.65 0.73 0.77 0.69 0.81 1.00 SM-25(1) 0.50 0.42 0.65 0.58 0.39 0.58 0.42 0.58 0.69 0.54 0.65 0.77 1.00 SM-7(1) 0.58 0.50 0.65 0.65 0.46 0.65 0.50 0.65 0.85 0.54 0.65 0.77 0.77 1.00 Similarity matrix was generated using value of simple matching coefficient (Ssm) and algorithm UPGMA.
I Wayan Suardana et al./Asian Pac J Trop Biomed 2015; 5(11): 915–920
transferred to humans through beef or contaminated environ-ment[3,5,6]. Other researches also reportedE. coliO157:H7 as
one strain of pathogenic E. coli known as a zoonotic agent could be detected in animals or humans [44,45] and the transmission usually associated with a complex factors involved environmental-host ecology that directly affects the likelihood of enterohemorrhagicE. coliO157[8,46,47].
Zoonotic potency of E. coli O157:H7 local isolates was supported by closely contact between human and cattle. In the area of study, cattle were not always cared by farmers in their cage, but also the farmer brought them outside, and cattle got feed or drinking water from their environment. On the other hand, the beef sold in traditional market was generally with poor hygiene so that the bacteria could be transmitted from animals to humans, according to the statement of previously researchers
[48,49]. In conclusion, the high degree of similarity of E. coli O157:H7 strains between isolates originated from cattle and human indicated the high potency of zoonoses, and AP-PCR method was proved to be a simple and rapid method to iden-tify the zoonoses agent ofE. coliO157:H7.
Conflict of interest statement
We declare that we have no conflict of interest.
Acknowledgments
The authors would like thank Prof. Dr. Supar MS for his kindness to give control isolates of E. coli O157:H7 ATCC 43894. The authors also wish to thank the Directorate of Research and Community Services, Directorate General of Higher Education which funded this research project through Udayana Research Grants with contract No.21.34/UN14/SBRC/ 2012, January 19th, 2012.
References
[1] Karmali MA, Gannon V, Sargeant JM. Verocytotoxin-producing
Escherichia coli(VTEC).Vet Microbiol2010;140(3–4): 360-70.
[2] Ateba CN, Mbewe M. Genotypic characterization ofEscherichia
coli O157:H7 isolates from different sources in the north-west
province, South Africa, using enterobacterial repetitive intergenic
consensus PCR analysis.Int J Mol Sci2014;15(6): 9735-47.
[3] Gallagher L, Soyemi K, Conover C, Austin C, Saathoff-Huber L,
Nelson S, et al. Outbreak ofEscherichia coliO157:H7 in a child
care center in Cook County, Illinois, with prolonged shedding and
household transmission.Am J Infect Control2013;41(10): 936-8.
[4] Amisano G, Fornasero S, Migliaretti G, Caramello S, Tarasco V,
Savino F. DiarrheagenicEscherichia coliin acute gastroenteritis in
infants in north-west Italy.New Microbiol2011;34(1): 45-51.
[5] Ferreira MR, Freitas Filho EG, Pinto JF, Dias M, Moreira CN.
Isolation, prevalence, and risk factors for infection by shiga
toxin-producing Escherichia coli (STEC) in dairy cattle. Trop Anim
Health Prod2014;46(4): 635-9.
[6] Brusa V, Aliverti V, Aliverti F, Ortega EE, de la Torre JH,
Linares LH, et al. Shiga toxin-producingEscherichia coliin beef
retail markets from Argentina.Front Cell Infect Microbiol2013;2:
171.
[7] Ateba CN, Mbewe M. Detection of Escherichia coli O157:H7
virulence genes in isolates from beef, pork, water, human and animal species in the northwest province, south Africa: public
health implications.Res Microbiol2011;162(3): 240-8.
[8] Ayaz ND, Gencay YE, Erol I. Prevalence and molecular
charac-terization of sorbitol fermenting and non-fermentingEscherichia
coliO157:H7(+)/H7(-) isolated from cattle at slaughterhouse and
slaughterhouse wastewater.Int J Food Microbiol2014;174: 31-8.
[9] Ateba CN, Mbewe M. Determination of the genetic similarities of
fingerprints fromEscherichia coliO157:H7 isolated from different
sources in the north-west province, South Africa using ISR,
BOXAIR and REP-PCR analysis. Microbiol Res 2013; 168(7):
438-46.
[10] Castro-Rosas J, Cerna-Cort´es JF, M´endez-Reyes E,
Lopez-Hernandez D, G´omez-Aldapa CA, Estrada-Garcia T. Presence of
faecal coliforms,Escherichia coliand diarrheagenicE. coli
path-otypes in ready-to-eat salads, from an area where crops are irrigated
with untreated sewage water.Int J Food Microbiol2012;156(2):
176-80.
[11] Page AV, Liles WC. Enterohemorrhagic Escherichia coli
in-fections and the hemolytic-uremic syndrome.Med Clin North Am
2013;97(4): 681-95.
[12] Ardissino G, Possenti I, Salardi S, Tel F, Colombo E, Testa S, et al.
Co-infection in children with bloody diarrhea caused by Shiga
toxin-producing Escherichia coli: data of the North Italian HUS
Network.J Pediatr Gastroenterol Nutr2014;59(2): 218-20.
[13] Bogard AK, Fuller CC, Radke V, Selman CA, Smith KE. Ground
beef handling and cooking practices in restaurants in eight states.
J Food Prot2013;76(12): 2132-40.
[14] Drastini Y. Identification and characterization based on genotype
and phenotype of verocytotoxigenicEscherichia coli(VTEC) from
livestock in Yogyakarta [dissertation]. Yogyakarta: Gadjah Mada;
2007.
[15] Suardana IW, Sumiarto B, Lukman DW. [Isolation and identifi
-cation ofEscherichia coliO157:H7 on beef at Badung Regency,
province of Bali].J Vet2007;8(1): 16-23. Indonesian.
[16] Suardana IW, Krisna Erawan IGM, Sumiarto B, Lukman DW.
Detection of Stx-1 and Stx-2 toxins ofEscherichia coliO157:H7
local isolates isolated from feces cattle and beef.J Vet2009;10(4):
189-93.
[17] Suardana IW, Artama WT, Asmara W, Daryono BS. Adherence
pheno-genotypic ofEscherichia coliO157:H7 isolated from beef,
feces of cattle, chicken and human.Indones J Biotechnol2011;
16(1): 46-52.
[18] Kudva IT.In vitroadherence patterns of Shigella serogroups to
bovine recto-anal junction squamous epithelial (RSE) cells are
similar to those ofEscherichia coliO157.Foodborne Pathog Dis
2012;9(4): 346-51.
[19] Glick BR, Pasternak JJ, Patten CL. Molecular biotechnology:
principles and applications of recombinant DNA. 4th ed.
Wash-ington, DC: ASM Press; 2009.
[20] Jaros P, Dufour M, Gilpin B, Freeman MM, Ribot EM. PFGE
for Shiga toxin-producingEscherichia coliO157:H7 (STEC O157)
and non-O157 STEC.Methods Mol Biol2015;1301: 171-89.
[21] Chiang YC, Lai CH, Lin CW, Chang CY, Tsen HY. Improvement
of strain discrimination by combination of superantigen profiles,
PFGE, and RAPD forStaphylococcus aureusisolates from clinical
samples and food-poisoning cases.Foodborne Pathog Dis2014;
11(6): 468-77.
[22] Pavlic M, Griffiths MW. Principles, applications, and limitations of
automated ribotyping as a rapid method in food safety.Foodborne
Pathog Dis2009;6(9): 1047-55.
[23] Williams JG, Kubelik AR, Livak KJ, Rafalski JA, Tingey SV.
DNA polymorphisms amplified by arbitrary primers are useful as
genetic markers.Nucleic Acids Res1990;18(22): 6531-5.
[24] Lee J, Kwon GH, Park JY, Park CS, Kwon DY, Lim J, et al.
A RAPD-PCR method for the rapid detection ofBacillus cereus.
J Microbiol Biotechnol2011;21(3): 274-6.
[25] Suardana IW, Artama WT, Widiasih DA, Mahardika IGNK.
Ge-netic diversity ofEscherichia coliO157:H7 strains using random
amplified polymorphic DNA (RAPD).Int Res J Microbiol2013;
4: 72-8.
[26] Moore JE, Watabe M, Millar BC, McMahon MA, McDowell DA, Rooney RJ, et al. Molecular characterization of verocytotoxigenic
Escherichia coli O157:H7 isolates by random amplification of
polymorphic DNA (RAPD) typing.Br J Biomed Sci2008;65(3):
161-3.
[27] Gamboa F, Chaves M, Valdivieso C. Genotypic profiles by
AP-PCR ofStreptococcus mutansin caries-active and caries-free
pre-schoolers.Acta Odontol Latinoam2010;23(2): 143-9.
[28] Akhi MT, Aghazadeh M, Kary NE, Darabi M, Barhagi MHS,
Pirzadeh T. Arbitrarily primed-polymerase chain reaction
(AP-PCR) typing ofClostridium difficileisolated from different sources
of Imam Reza hospital, Tabriz, Iran.Afr J Microbiol Res2013;
7(21): 2475-80.
[29] Barone S, Macedo C, Marin JM. Arbitrarily primed polymerase
chain reaction for fingerprinting the genotype identification of
mutans streptococciin children with down syndrome.Spec Care
Dent2005;25(1): 37-42.
[30] Wangpermtam P, Sanguansin S, Petmitr S, Punyarit P,
Weerapradist W. Genetic alteration in oral squamous cell carci-noma detected by arbitrarily primed polymerase chain reaction.
Asian Pac J Cancer Prev2011;12(8): 2081-5.
[31] Jiang Q, Yu M, Min Z, Yi A, Chen D, Zhang Q. AP-PCR detection
ofStreptococcus mutansandStreptococcus sobrinusin caries-free
and caries-active subjects.Mol Cell Biochem2012;365(1–2):
159-64.
[32] Chuensumran U, Saelee P, Wongkham S, Pairojkul C, Chauin S,
Petmitr S. Histological type of intrahepatic cholangiocarcinoma
differentiated by genetic alteration from AP-PCRfingerprint.Asian
Pac J Cancer Prev2011;12(6): 1377-80.
[33] Hosbul T, Ozyurt M, Karademir F, S ¨uleymanoglu S,
Haznedaroglu T. [Investigation of a nosocomial outbreak caused
by ESBL positiveKlebsiella pneumoniaein neonatal intensive care
unit by AP-PCR].Mikrobiyol Bul2012;46(1): 101-5. Turkish.
[34] Punina NV, Zotov VS, Parkhomenko AL, Parkhomenko TU,
Topunov AF. Genetic diversity of Bacillus thuringiensis from
different geo-ecological regions of Ukraine by analyzing the 16S
rRNA and gyrB genes and by AP-PCR and saAFLP.Acta Naturae
2013;5(1): 90-100.
[35] Suardana IW, Pinatih KJP, Ratnawati NLKA, Widiasih DA.
Protein profile analysis of Escherichia coli O157:H7 from
human and animals origin.Int J Curr Microbiol Appl Sci2013;
2: 204-14.
[36] Suardana IW. Analysis of nucleotide sequences of the 16S rRNA gene of novelEscherichia colistrains isolated from feces of human and Bali cattle. J Nucleic Acids 2014;http://dx.doi.org/10.1155/
2014/475754.
[37] Welsh J, McClelland M. Fingerprinting genomes using PCR with
arbitrary primers.Nucleic Acids Res1990;18(24): 7213-8.
[38] Al-Darahi KF, Mahdi LK, Al-Naib KT, Jubreal J. Molecular
characterization ofE. coliO157:H7 strains using random amplified
polymorphic DNA (RAPD).J Dohuk Univ2008;11(1): 198-204.
[39] Atri M, Nematian MA, Shahgolzari M. Determination and
discrimination of intraspecific diversity ofAstragalus gossypinus
by eco-phytosociological method from west of Iran.Pak J Biol Sci
2007;10(12): 1947-55.
[40] Dam´e-Teixeira N, Arthur RA, Parolo CCF, Maltz M. Genotypic diversity and virulence traits ofStreptococcus mutansisolated from carious dentin after partial caries removal and sealing.Sci World J
2014;http://dx.doi.org/10.1155/2014/165201.
[41] Madico G, Akopyants NS, Berg DE. Arbitrarily primed PCR DNA
fingerprinting ofEscherichia coliO157:H7 strains by using
tem-plates from boiled cultures.J Clin Microbiol1995;33(6): 1534-6.
[42] Ranjbar R, Pourshafie MR, Sadeghifard N, Karami A,
Hamidian M, Izadi M, et al. Molecular characterization of epidemic
isolates ofVibrio cholerae O1 by arbitrarily primed PCR
(AP-PCR).Iran J Public Health2008;37(2): 83-7.
[43] Rossell ´o-Mora R, Amann R. The species concept for prokaryotes.
FEMS Microbiol Rev2001;25(1): 39-67.
[44] Krauss H, Schieffer HG, Slenczka W, Weber A, Zahner H.
Zoo-noses: infectious diseases transmissible from animals to humans.
3rd ed. Washington, DC: ASM Press; 2003.
[45] Launders N, Locking ME, Hanson M, Willshaw G, Charlett A, Salmon R, et al. A large Great Britain-wide outbreak of STEC O157 phage type 8 linked to handling of raw leeks and potatoes.
Epidemiol Infect 2015; http://dx.doi.org/10.1017/S095026881
5001016.
[46] García A, Fox JG, Besser TE. Zoonotic enterohemorrhagic
Escherichia coli: a one health perspective. ILAR J2010; 51(3):
221-32.
[47] Beutin L, Martin A. Outbreak of Shiga toxin-producing
Escher-ichia coli(STEC) O104:H4 infection in Germany causes a para-digm shift with regard to human pathogenicity of STEC strains.
J Food Prot2012;75(2): 408-18.
[48] Gaulin C, Ramsay D, Catford A, Bekal S.Escherichia coliO157:
H7 outbreak associated with the consumption of beef and veal
tartares in the province of Quebec, Canada, in 2013.Foodborne
Pathog Dis2015;12(7): 612-8.
[49] Baba A, Ebuchi S, Uryu K, Hiwaki H, Ogata K, Washimi E, et al.
An outbreak of water-borne gastroenteritis caused by diarrheagenic
Escherichia colipossessing eae gene.Jpn J Infect Dis2006;59(1):
59-60.
I Wayan Suardana et al./Asian Pac J Trop Biomed 2015; 5(11): 915–920