Production of Patient Specific
Probes
for
the Detection of
Minimal
Residual
Disease
in
Acute
Lymphoblastic
Leukemia
Dian
Tirza",
Marleen Bâkkus*"
Abstrak
Dua puluh dua penderita ALL (acute lymphoblostic leukemia) pada anak-analç setelah didiagnosis Lalu dianalisis konfigurasi Ig (imtnunoglobulin) dan TcR-6 (T-cell receptor-delra) gennya dengan ,netodc Southern blot. Pendetel<sian gene rearrangements ini
menunjukkan apaknh junctional region lconfigurasi
IgH
(immunoglobulin heavy chain) atau TcR-6 akan dianalisis dengan PCR (polymerase chainreaction). UrutannuHeotidaJunctional regions dari rearrangedlgH (inmunoglobulinheavy chain) danTcR-6 genes ditentukan dengan direcr sequencing hasil produk PCR daerah ini. Berdasarkan data sekuens ini, nnka pelacakyang spesifik terhadap pasien tersebut (patient-specific oligonucleotide probes) akan didesain. Selanjutnya, su,nsu,n tulang dan darah tepi yang diambil selann dan setelah pengobatan, akan dianalisis dengan teknik PCR nenggunal<an pelacak yang spesifik terhadap pasien tersebut untuk nendetelrsi sel leukenik Linn pelacakyang spesifik telah didesain dan diuji spesifisitos dan sensitivitasnya.Abstract
Twenty-two patients with childhood ALL were analyzed
for
the configuration of theirIg (innunoglobulin) and
TcR-6 (T cell receptor delta) genes by Southern blot analysis- The detection ofgene rearrangenents indicatedwhether IgH (itnmunoglobulin heavy chain)or
TcR-6 junctional regiotts should be analyzed by PCR (Polymerase chain reaction). The nucleotide sequence ofjunctionalregions of the rearranged IgH and TcR-ô genes are deternined by direct sequencing of PCR products of these regiotrs. Based on the sequence data, patient-specific oligonucleotide probes were designed- Subsequently, bone narrow
and/
or peripheral blood sanples taken daring andafier treattnent,will be analyzedwiththe PCRtechnique, using patient - specific oligonucleotide probesfor the detectionofleukenic cells. Five patient specffic probes have been designed and tested in specificity and sensitivity.
Keywords : ALL (Acure L),nphoblastic Leukenia), MRD (Mininal Residual Disease),
IgH
(lnnunoglobulin Heavy Chain) gene, Tc R-6 (T- c ell r ec epto r -d eha) gene.INTRODUCTION
Approximately
20-30
%of children
with
ALL
showeda relapses,
in
spiteof major improvements in the
treat-ment achievedduring
the lasttwo
decades.Apparent-ly,
the current treatment is
not
adequateto
kill
all
theleukemic cells, although
the vast
majority
appear
toreach
complete
remission
according
to
the
current
morphological detection technique.
The
detection
limit of
this
technique
is
about
I-5%
(L-5
leukemic
cells
in
100normal
cells).
It
is obvious that this
tech-nique
only
provides superficial information
on
theresult
of
leukemia
treatment."'''
Therefore, more sensitive
techniques arerequired
for
the
detection
of lower
numbers
of
leukemic cells
to determine whether the tumor load can be decreasedduring
treatment,The
terminology of MRD (minimal
residual
dis-ease) means that the leukemic cells are present
in
theperipheral
blood
or
bone
below
thedetection
limit of
conven
Now
by using recombinant
DNA
technology
it
ispossible
to
recognize
MRD in
ALL
by
analyzing
the rearrangementsof
theIg
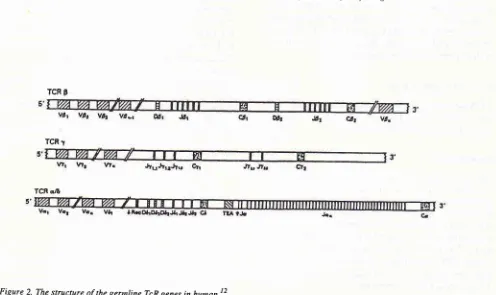
and TcR genes, asillustrated
in Figure
I
and Figure 2.* In partial fulfillntent of the requirenenîs
for
the degree of Moster of Science in Molecular Biology, Vrije Universiteit Brussel (V.U.B), Belgium.*
Deparnnent of Pharnacologl,, Facultl, of Medicine University of Indonesia, Jakarta, Indonesia.Vol 3, No 3, July - September 1994
The
IgH
genes arerearranged
:rr^98%ofcases
of
B-cell
ALL,
and 45-50%
rearranged
Ig
light
chain
(IgL)
genes.5'6'?'8 Ther""rr".g"-"rrt
of IgHlenes
aredetectable
with
a
JH
probe
(seeAppendix
l)
after
digestion
with
therestriction
enzymesof Bglll,
BamHI,
andHindlll.
The
rearrangementsof
TcR-ô
genesoccur
in
more than 90%
of T-ALL,
65%-70%
of
precursor
B-ALL
which
is
detectable
by
Jôl
probe
(see
Appendix
1).The
general
aim
of
the
project
was detection
of
MRD in
childhood of
ALL,
by studying
thejunctional
region
of IgH
andTcR-ô
gene rearrangementsby
thePCR. Therefore,
first of all
the
configuration
of
their
IgH
and TcR-ô
genes
were
analysed
by
use
of
Southernblot.
Subsequently, the sequenceofjunction-al regions
of
rearrangedIgH
andTcR-ô
geneswill
bedetermined
by direct
sequencing
of
PCR products of
these
regions.
From the
sequencedata
can
produce
patient
specific
probes.
Finally, this information
will
be
used
to
detect
leukemic cells after
treatment
andevaluate
whether the
treatment
of ALL
can be
im-proved.
MATERIALS
AND METHODS
Cases
The
biological
specimens that was studied consistedof
peripheral blood (PB)
and bonemarrow
(BM)
samplesobtained
from 22
children
with immunophenotypic
common
ALL, B-cell ALL
andT- cell
ALL.
They aged between2 and9
yearsold,
Thesechildren
were treatedwith different multidrug
regimens.I
solation of
ly mp h o cy te sDNA
was
extracted
from fresh cells
or
frozen
cells
which
were isolated
from
PB
or BM
according to
the method describedby
Maniatis
et al,eSouthern
Blot
Ten ug
of this DNA
wasdigested
with
therestriction
enzymes (Pharmacia, Uppsala, Sweden),
electro-phoresedn
O.8% agarosegel,
andtransferred onto
aHybond
N* membrane
(Amersham, Tokyo,
Japan).Then hybridization
of
Southern
blots
consisted
of
prehybr
with
r
washes.
the sizes
of
germline and
rearrangement
restriction
fragments
in Kb
is aprerequisite
for
an accurateresult
(seeAppendix
2).Production of Patient-specific
Probes
149Poly merase
chain reaction
PCR was carried out as described
by Saiki
etallo using
the denaturation step at 94oCfor
3O seconds, ananneal-ing
step
at
55oCfor
30
seconds,and
anextension
at72"C
for
I
minute
20
seconds.This
was done
for
40cycles
(for IgH).
For Vô2-Dô3
rearrangements,
the sampleswere subjected
to
the
following cycles
:
thedenaturation
step at95"C
for
5minutes,
theannealing
step at 66oCfor
2 minutes,
andthe extension
at72oC
for
4O seconds.This
was done
for 40
cycles
in
Ther-mocycler (Bio-Med, B.Braun, F.R.G).
As negative
control
sample,
a reaction
was
performed
with
HzO insteadof
template
DNA.
Oligomers
usedin
thePCR
is
describedin Appendix
3.Direct
sequencing
Direct
sequencing
step
consisted
of
purification
of
amplified
DNA
(Promega
Corporation,
USA),
pqe-paration
of
DynabeadsM-280 streptavidin
lDynai(R),
Norway
CoAs),
separationof
DNA
strands, annealingmixture,
labeling reaction
andtermination reaction.
The
biotinylated primer
was
usedfor
direct
sequenc-ing.
Dot
Blot Hybridization
The
PCR
product
was
serially
diluted loo-loa
times
starting from 5
pl
(1:10
of
the total volume
of
PCRproduct),
then denaturatedby adding
4 prlof 5N NaOH
+
I
Fl
of
0.5M EDTA (pH
8.2) andincubated
at 950Cfor
l0 minutes. The
sampleswere then
chilled
on ice
and
50
pl of
2N
NH4
acetate
pH
7.O was
added.Subsequently,
the samples were immobilized
on a
piece
of
Zeta
Probe
Membrane (Biorad
membranesNo:
162-0153),
using the Bio-rad dot blot
apparatus(Biorad). After application
of
the
DNA
on the
mem-brane, the wells were rinsed
with
500
pl of
0,4 N
NaOH.
After removing
the
membrane,it
was washedin 2X
SSCfor
5minutes (3 times).
Subsequently, the membranewas
air
dried
andstored
or hybridized
im-mediately,
RESULTS
Configuration
of
the
Southern
blot
TcR-ô
and IgH
genes by
gerrnline lgH genes
vtt
123456
n1234
0nJHsCp
123456
D to J joining
prècursor lgH mRNA
iYi
I
+
RNA splicing mature lgH mRNA [image:3.595.50.542.70.414.2] [image:3.595.45.541.439.734.2]VDJ
GpFigure 1' Rearrangeuent ofinnunoglobulin heavy chain gene. First D to J
ioining
occurs,followed by V to D-J joining. The rearranged genes can be transcribed into a precursorIgH
tRNA; which becomes o ,roturcIgH
ntRlrilafier
spl'icing.l
t'12'
--'
-'-cTCR 9
TCR
r
TCR qzE
Vol 3, No 3, July - September 1994
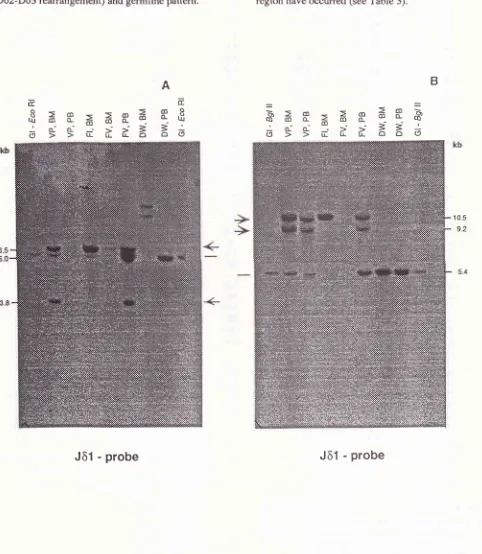
the
Jôl
prob.
BM
and / or PBDNA
of
4ALL
sampleswas cut
with
EcoRI
(Figure 3A)
or BgZII
(Figure 3B).
ln
BgL
II
andEco RI,
these samples showed rearran-gementsof
theTcR-ô
gene(Vô2- Dô3
rearrangement,Dô2-Dô3 rearrangement)
andgermline
pattern.ProductionofPatient-specific
Probes
151Twenty two
ALL
patients were
analyzedby
Southernblotting.
The results areshown in
Table
l.
Analysis
of
all
thesedata
indicated that
rearrangement
of
TcR-ô
region
(seeTable
2)
andrearrangement
of
IgH
generegion
haveoccurred
(seeTable
3).105
92
B
A
Ecc
,fËBËËæaeÊ
6ee;'ùÈââà
==
R=R=;æâæR
Éee;'eÈââ6
[image:4.595.54.536.144.698.2]Jô1
- probe
Jô1 - probe
Figure 3. TcR-6 gene configuration in 4 ALL patients. DNA santples
fron
Blr4/or PB santples were digested with EcoN
(A) and BglII
Table 1. Configuration of IgH and TcR-ô genes rearrangements in childhood ALL by Southem blot analysis. Name of
patients
Source* Eco Rl IgHBglll
Eco RITcRô
BslIJBF BN BN BC BC
cw
cw
CK CK DB DB DC DW DW FV FV FI GI GL HT HB HB JAKY
KY
LN LN RM RM VB VB VC VC VU VU VS VS VP VP BM BM PB BM PB BM PB BM PB BM PB BM BM PB BM PB BM BM PB PB BM PB BM BM PB BM PB BM PB BM PB BM PB BM PB BM PB BM PB R**
NR NR NR ND ND ND ND G 2R R 2R 2R R ND ND NR R ND 2R ND ND G ND NR R ND R NR G R ND 2R R D/R R R G NR NR ND ND ND ND G 2R R 2R 2R D/R ND ND NR R ND 2R ND ND G/R ND NR G/R R R G/R G/R R ND 2R R R R D/R(Vô2-Dô3) R(Vô2-Dô3) G R(Vô2-Dô3) R(Vô2-Dô3) ND G ND ND R(Dô2-Dô3) ND R(Vô2-Dô3)2R* (unknown) G
R(Vô2-Dô3) G/2R (Vô2-Dô3, Dô2-Dô3) G/R(Vô2-Dô3) ND ND NR R(vô2-Dô3) ND 2R(Vô2-Dô3, Dô2-Dô3) ND ND G G G G ND ND R(Vô2-Dô3) NR R(Vô2-Dô3) ND R(vô2-Dô3) R(vô2-Dô3) G/2R(Vô2-Dô3, Dô2-Dô3) NR D/R(Vô2-Dô3) R(Vô2-Dô3) G R(Vô2-Dô3) R(Vô2-Dô3) ND G ND ND R(Dô2-Dô3) ND R(Dô2-Dô3) G G NR
G/2R (Vô2-Dô3, Dô2-Dô3) R(vô2-Dô3) ND ND NR R(Vô2-Dô3) ND 2R(Vô2-Dô3, Dô2-Dô3) ND ND G G G G ND ND R(Vô2-Dô3) NR R(vô2-Dô3) ND R(Vô2-Dô3) R(Vô2-Dô3) R(vô2-Dô3) G/2R(Vô2-Dô3, Dô2-Dô3) Abbreviation:
NR : no result due to technical problem.
R
: gene segment in rearrangement conhguration of Jôl.
ND : not determined because not enough sample.
G
: gene segmen in germline conhguration of Iô1.D
: deletion ofthe involved gene (segment). [image:5.595.78.570.102.661.2]Vol 3, No 3, July - September 1994
Table 2. TcR-ô gene rearrangements in childhood ALL. T cell
receptordelta
Eco RI BglrtReânângemenls of TcR-ô
One allele rearranged Both alleles rearranged One allele deleted /
one rearranged
t4t22 (63.63%)- 14122 (63.63%)
Number of cases demonstrating reanangements / numbr of cases
studied.
Calculation data were obtained from table 1.
Table 3. Immunoglobin gene reârrangements in childhood ALL.** Immunoglobulin
Heavy
EcoRI
Bglll
chain
Reanangenrents of IgH genes
One allele reananged Both alleles rearranged
One allele deleted /
one tearranged
t4122 (63.63%)* 15122 (68.r8%)*
* Number of cases demonstrating rearrangements / number of cases
studied
** Calculation data were obtained from table l.
The detection of
IgII
and
TcR-ô
rearrangements
in
childhood
ALL
by PCR technique
Most
of
the
ALL
patients had a
Vô2-Dô3
rearrange-ment
asjudged by
Southern
blot
analysis. Therefore,
first
of
all
the Vô2-Dô3
junctional
region was
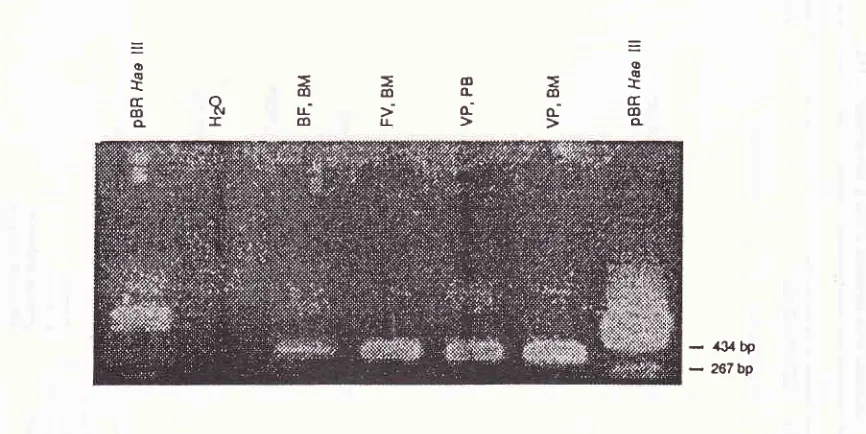
ampli-fied. The amplification
products were
separatedin
aProductionofPailent-specific
Probes
153L5%
agaræegel
(Figure
4).
The
sizesof
thesejunc-tional
region
were 400 bp.The amplification
of
the IgH junctional
region
was
performed by PCR using
theprimers
FR3-5'
andLJH-3'.
The
sizesof
thejunctional
regions were
be-tween
180-200 bp.Fourteen
casesout of the
22
ALL
patients
(63.64%) indicated
amplification
of Vô2-Dô3
rearran-gementsby
PCR
technique.
Eight
casesdid
notshow
Vô2-Dô3 rearrangements.
Seventeen
cases
out
of
the
22
ALL
patients
(77.27%) gave
amplification
products
when
the
IgH
junctional
regions were
amplified.
In 4
casesno
bandwas
obtained.
One case wasnot yet
analyzed.The nucleotide
sequences
of
junctional
regions
of
rearranged TcR-ô
genes.The
14 casesof
ALL
with a
Vô2-Dô3
rearrangementwere
chosen
for
direct
sequencing.
The biotinylated
primer
was used
for
direct
sequencing
after
gel
purification.
The
PCRproducts were
sequenced. Thesequence data
were
comparedto published
sequencesconfirming
that
thePCR products derived
from
Vô2-Dô3 genes
rearrangements as
shown
in
table
4.
Se-quences
which are identical
to Vô,
Dô and
Jôl
germline
sequences wereidentified. Other
nucleotides
in this region were
judged
a N region.In
7
of the
14patients unique
sequences areobtained.
In
5 cases nomonoclonal
sequence
was
obtained.
Two
casesshowed unreadable
sequences.
The
sequences
of
patients
KY
andVP
showed adeletion
of
Dô3
gene. rol223122
tl22 tol22
3122
u22
t0122
4122
u22
9122 4122
tl22
cÛd)(L
rr-'to.
(D TI
=
o6
I
(rO
(I)
sl
CLI
-
€1
bp [image:6.595.58.293.100.378.2]-
2ATbp [image:6.595.96.529.497.714.2]É
ËË
9 èoË.EEe
ri
Ei':r-ËËË€g
o: o o d-Ëao!o
!
..I:
B.EFË
Ë=ooo a
àEEE
85€€€Ë!
3 é É é'â
<)))È.Ë
ôu
3
I
UId
Fo
O C)e
(, (, (, () (Jo
F () L Uo
t-(, ll(l
d
UI H (J,
t-..c)o
vttib!
3Ég
EEP
F FE8
OO(,()
t-
Vol 3, No 3, July - September 1994
Dot blot hybridization
Five patient specific probes
were used tohybridize
theDNA
samples, except
2
patient
specific
probe.s(VP
and
JA).
To
test the specificity
of
patient speg$c
oligo's,
theoligo's
were endlabeled
with
gamma-t'P-dCTP's
and used
in
a dot blot hybridization
experi-ment.Production of Patienr-specific
Probes
155The
oligo's only hybridized
to
theDNA
samplesfrom
which they derived (Figure
5).
The
detection
sensitivity
wasvariable.
ProbesHB, VB
andBC
only
hybridized weakly probably
due
to the
length
of
theoligo
andconsequent\
thelow
hybridization
tempera-ture(32"C).
ProbesKY
andBN
gave a strongersignal,
both
theseprobes
had aTm
(temperature
melting)
of
420C.
A
: the membrane was hybridized to probe KY. B : the membrane was hybridized to probe BN. C : the membrane was hybridized to probe HB.D : the membrane was hybridized to probe VB.
E : the membrane was hybridized to probe BC.
The samples were diluted stârting from 5 pl of amplification DNA in ten fold serial dilution.
123,1
5I
6 7 I 9 10
11D
3 4 5 6 7 A 9 rO ll
E
t23_,t567891011
I
,Ft4,zz:eJ.2zz2tz:a/22Z2,l2.tzztzV/ztzaratzV/t72eàà7PC.?(/à2-DJ),
probe. Arrow indicates the detection of leukemic cells.
roo to't
rc-2
Lane
I
: normal control DNA.Lane 2:
patient VB.Lane 3:patientHB.
Lane 4:
patient BC.Lane 5:patientBN.
Lane 6:patientKY.
Lane 7:patientVS.
Lane 8:patientV4
Lane 9:patientVP
Lane 10 : patient DB
DISCUSSION
Most
of
the
ALL
patients have
TcR Vô2-Dô3
re-arrangementsand
IgH
rearrangement
as
detectedby
Southern
blot. Therefore, in
most casesit
is notneces-sary
to perform
Southern
blot
analysis,
but only
per-form
the PCRwith
theVô2-5'/Dô3-3'
primers
and rheFR3/LJH
primers.
Only
in
the
case
of
negativity
Southern
blot
can beuseful
tojudge
theTcR
andIgH
rearrangement.We set up a PCR assay based on the
variability
of
the
IgH
andTcR-ô
genein
ALL.
This PCR technique
allows the
detection
of
MRD in
ALL.
By
PCR,
thejunctional region
of
TcR-ô
rearrangement
could
beamplified
in
14 casesout
of
22
(63.64%).
Seventeencases
out
of
22 (77.27%)
gaveamplification
product
of
the IgH
rearrangement. One
of
the
advantagesof
PCR are that the technique requires lessDNA
than theSouthern
blot
technique.
It
is
more sensitive
and lesstime consuming
in
comparison
with
the Southemblot
technique. The probes
specific for
thejunctional
region
of the
TcR-ô region
are now used in thefollow
upstudy
these patients.
Whether
thedetection of
MRD
by PCR
technology
will
lead to
decreaseof
themorbidity
andmortality
in
ALL
can
only be
answered
after
multi-centre
evaluation
of
the technique.REFERENCES
l.
Katz F, Ball L, Gibson B, Chessels J. The use of DNA probes to monitor minimal residual disease in childhood acutelym-phoblastic leukemia. Brit J Haematol 1989;73:173-80. 2. Fey MF, Wainscoat J. Molecular diagnosis of hematological
neoplasms. Blood reviews 1988;2:78-87.
3. Jonsson OG, Kitchens RL, Baer RJ, Buchanan GR, Smith.
Rearrangements of the tal-1 l.ocus as Clonal Markers for T
Cell
Acute Lymphoblastic
Leukemia.J
Clin
InvestI99I;87:2029-35.
4. Potter
MN.
The detectionof
minimal residual disease inacute lymphoblastic leukemia. Blood reviews 1992;6:68-82.
5. Toyonaga
B,
YoshikaiY,
VadaszV,
ChinB,
Mak TW. Organization and sequencesof
the diversity, joining, andconstant region genes
of
the human T-cell receptor Beta chain. Proc Natl Acad Sci USA 1985;82:8624-8.6. Korsmeyer SJ. Antigen receptor genes as molcular markers of lymphoid neoplasms. J Clin Invest t987;79:L29L-5. 7, Nadler
LM,
Korsmeyer SJ, AndersonKC, Boyd
AW,Slaughenhoupt
B,
Park E, JensenI,
Conirl F, Mayer RI,Sallan SE, Ritz I, Schlossmann SF. B cell origin of non T-cell acute lymphoblastic leukemia.
A
model for discrete stagesof neoplastic and normal pre-B cell differentiation.
I
ClinInvest 1984;74 :332-4O.
8. Foa R, Migone N, Basso G, Cattoretti G, Pizzolo G, Lauria,
F,
CasoratiG,
GiubellinoMC,
CapuzzoF,
Rajnoldi C,Lusso, P, Carbonara
AO,
Gavosto F. Molecular andim-mnological evidence of B-cell commitment
in
"Nul" acutelymphoblastic leukemia. Int.
f
Cancer 1986;38:317-23. 9. Maniatis T, Fritsch EF, Sambrook J, In: Molecular CloningA Laboratory Manual. Cold Spring Harbor, New York,1982. 10. Saiki RK, Gelfand DH, Stoffel S, Scharf SJ, Higuchi R, Horn
GT,
Mullis KT,
Erlich
HA.
Primer-directed enzymatic amplification of DNA with termostable DNA polymerase. Science 1988 ;239:487-9l.
11. Van Dongen JJM, Wolvers-Tettero
ILM.
Analysisof
im-munoglobulin andT
cell receptor genes. Clinica Chimica Acta 1991;198:1-92.12, Potter MN. The detection
of
minimal residual disease inacute lymphoblastic leukemia. Blood reviews 1992;6:68-82.
13.
Felix CA,
PoplackDG.
Characterizationof
acutelym-phoblastic leukemia of childhood by immunoglobulin and T
cell receptor gene patterns. Leukemia 1991;5:1015-25. 14. Quetermous T, Strauss WM, Van Dongen JJM, Seidman JG.
Human
T-cell
gamma chainjoining
regions and T-cell development. J of Immunolo gy L987;8:2687 -9O.15. Jong D, Voetdijk BMH, Van Ommen GJB, Kluin PM.
Al-terations
in
immunoglobulin genes reveal the origin andevolution
of
monotypic andbitypic
B
cell
lymphomas. American Journal of Pathology 1987 ;134:1233-42.16. Breit TM, Wolvers-Tettero
ILM,
HahlenK,
Van Wering,ER, Van Dongen JJM. Extensive junctional diversity
of
Gamma-delta
T -
cell teceptors expressed by T-cell acutelymphoblastic leukemias : Implications for the detection
of
Vol 3, No 3, July - September 1994 Production of Patient-specific
Probes
I57
Appendix 1. The probes required
for
southern blot analysisof
Apeendix 2. Approximate sizesof
germline andrearrange-TcR.ô gene reanangements and IgH reanangement ment restriction fragments in kb (exon content
of
restriction fragment TcR-ô gene)
Eco
RI B9III
Hindlll
BamHl Kpnl
germline
Iôl
6.0
5.4 6.0 I7.5
17.5Vôr-Jôt
3.3
5.6 11.0
>30
9.4JH
probe Clone
: Pvu ll fragment of the JH genesegment
Vôz-Jôr
5.5
lO.2
6.1
20
8.4J ôl
probe Clone
: pJ ô516Vectror
: pUCResistance :Ampicillin
Insert (total) : Sac I- Sac I
-
1.5 kbBacteria
: DH5aF (11,14)Vector
: in pBR 322Resisoance :Ampicillin
Ins€rt(total) : BamHl-HindlII' 6 kb
Inset (probe) : Saa 3A I - 2.0 kb
Bacteria
: DHI (15)Vôz-Dôr
6.5 10.5 7.O 21
9.4Dôz-Dôg 3.8
9.2 5.8 11.0
6.7Dôz-Iôr
2.8
8.2 4.8 10.0
5.7Appendix 3. Oligomers used in the PCR and / or ditect sequencing analysis of TcRô and IgH genes reânangements
The name
ofcode
PCR/ Sequence SequenceTcR$
genesVô1-5'backward
Vôl-3'backward
Vô2-5'backward
Vô2-3'backward
Dô3-3' forward
Iô1-3'forward
PCR
PCR/Seq
PCR
PCR/Seq*
PCR
Seq
Xbal
GAAGATCTAGACTCAAG CCCAGTCATCAGTATCC
Bgln
Sall
l-{CGCGTCGACGCCTTAACCATTTCAGCCTTAC
Sall
CGCGTCGACCAAACAGTGCCTGTGTCAATAGG
CGCGTCGACCTGGCTGTACTTAAGATACTTGC
Sall
GTAGATCTAGAAATGGCACTTTIGCCCCTGCAG H
Bgln
GAAGATCTAGACCTCTTCCCAGGAGTCCTCC
Bsllt
IgH genes
FR3-5'backward
LIH-3'forward
PCR/Seq
PCR/Seq*
ATGGAATTCACACGGC(CTXGC)TGTATTACTGT Eco RI
CACCTGAGGAGACGGTGACC
5' and 3' extension of the nucleotide names indicates the location of the oligonucleotide primer within the gene segment. The restric-tion sites are indicated. Oligonucleotide primers used in PCR or sequence analysis are indicated
P
iR and sequence, respectively lÔ.