Characterization of a periclinal chimera variegated tobacco
(
Nicotiana tabacum
L.)
Chang-Hyu Bae
a, Tomoko Abe
a,*, Noriko Nagata
a, Nobuhisa Fukunishi
b,
Tomoki Matsuyama
a, Takeshi Nakano
a, Shigeo Yoshida
aaPlant Functions Laboratory,RIKEN(Institute of Physical and Chemical Research),2-1Hirosawa,Wako,Saitama351-0198,Japan bRI Beam Factory Project Office,RIKEN(Institute of Physical and Chemical Research),Wako351-0198,Japan
Received 12 April 1999; received in revised form 4 October 1999; accepted 5 October 1999
Abstract
A plant isolated from a population of tobacco (Nicotiana tabacumL. cv. BY-4) lines, which were generated with a heavy-ion beam, exhibited green and white variegation on its leaves, stems, and calyces. Fluorescence microscopic analysis of white and green sections of the leaves of the mutant showed an absence of developed chloroplasts in the epidermis and palisade layer. There were undeveloped chloroplasts in one or two layers of spongy parenchyma cells in the green sections of the mutant. In the F1
generation, the segregation ratio of green to white was 6:1163 and reciprocal crosses showed that the variegation was not maternally inherited. These results suggest that the mutant is a periclinal chimera with a White-White-Green histogenic composition. The mRNA levels of the photosynthetic genes rbcL andpsbA in the green areas were normal, while they were significantly reduced in the white areas. © 2000 Published by Elsevier Science Ireland Ltd. All rights reserved.
Keywords:Variegated tobacco; Periclinal chimera; Chloroplast gene
www.elsevier.com/locate/plantsci
1. Introduction
The classic studies of the non-Mendelian inheri-tance of green – white variegation [1] provided sup-port for the existence of extra-nuclear DNA. Recent studies have reported that this green – white variegation is associated with the alteration of chloroplast [2 – 5] or mitochondria [6 – 10] genes. Periclinal chimeras reveal that all three apical lineages contribute to the leaf primordium in to-bacco [11 – 13]. The cell lineages in periclinal chimeras are referred to as L1 (layer 1), L2 (layer 2), and L3 (layer 3). L1 contributes exclusively to the epidermis; L2 produces the subepidermal layer of the mesophyll near the center of the leaf; and L3 gives rise to the middle mesophyll layers in the center of the leaf blade [13]. These relationships
have been established in genetic and histological studies of tobacco [11], poinsettia, and carnation [14]. Periclinal chimeral tobacco plants can have G-G-G (L1-L2-L3), G-G-W, G-W-W, W-W-W, W-W-G, and G-W-G (G, green; W, white) histo-genic types in stable homoplastidic periclinal chimeras [12]. Of these six histogenic types, the leaf phenotypes and the modes of segregation of types G-W-G and W-W-G leaves are very similar, except that the epidermis is green or white, respec-tively [12].
Several well-known examples of variegation are inherited maternally [3,4,6 – 8,15,16]. This involves an inherited nuclear gene system [2,17] with a non-Mendelian inheritance pattern [9,18]. In addi-tion, various modes of inheritance have been re-ported for the leaf histological patterns seen in variegated tobacco plants [11 – 13]. For example, type W-W-W only produces white progeny, while types G-W-G and W-W-G produce very few green progeny by self-pollination.
* Corresponding author. Tel.:+81-48-467-9527; fax:+ 81-48-462-4674.
E-mail address:[email protected] (T. Abe)
An approach using leaf tissue that has naturally lost the ability to photosynthesize is ideal for studying the expression and regulation of genes related to photosynthesis [6,19]. Genes that di-rectly or secondarily affect chloroplast form and function have been identified in variegated mu-tants [2,3,8,10,16]. Therefore, the variegated areas of mutants are extremely useful for studying alter-ations of photosynthetic genes [7] and they are an excellent starting point to dissect the poorly under-stood pathways of communication between the nuclear-cytoplasmic, chloroplast, and mitochon-drial genetic systems [20,21]. In this study, we present the genetic, histological, and molecular characteristics of a variegated tobacco phenotype, which is a periclinal chimera that has undeveloped chloroplasts in some of the leaf mesophyll cell layers.
2. Materials and methods
2.1. Plant materials
The ovaries of tobacco (Nicotiana tabacum L. cv. BY-4) containing embryos 24 – 96 h after self-pollination were irradiated with a14N beam (135A
MeV) accelerated by the Riken Ring Cyclotron with a dose range of 5 – 200 gray. The linear energy transfer of the nitrogen ions corresponded to 28.5 keV/mm [22]. One month after irradiation,
seeds of the M1 generation were harvested. A
variegated plant was found in an M2 population
derived from self-pollinated M1 plants that had
been irradiated with 50 gray of a N-ion beam 84 h after pollination. The M2seeds were germinated in
half-strength Murashige and Skoog (MS) [23] medium containing 2.0% sucrose and 0.8% agar at 25°C under continuous light (80mmol quanta m−2
s−1). Two weeks later, the variegated plant was
transferred into a plant box to be grown under the same condition. After 2 months, the plant was planted in a plastic pot containing soil, and was placed in a greenhouse under 18-h photoperiod (100 mmol quanta m−2 s−1) at 25°C. A fully
expanded upper leaf of the plant was harvested monthly to be used in biochemical experiments. The variegated plant was permitted to carry flower at a 6 month-old stage to achieve for self-pollina-tion analysis.
2.2. Chlorophyll estimation
The chlorophyll content was determined using a modified version of the procedure of Aron [24]. To determine the chlorophyll content, 0.3 g (fresh weight) of leaves from white and green areas of a variegated plant were homogenized in 80% ace-tone and centrifuged at 9000×g for 10 min. The clear supernatant was then diluted to 1:80 in 80% (v/v) acetone, and the absorption spectra were determined at 645 and 663 nm.
2.3. Genetic analysis
The M2 variegated plant was derived from M1
line S201 [25]. In M2progeny of the S201
self-pol-lination, the segregation ratio among green, white and variegated was 2588:587:1 and reciprocal crosses of the S201 line with wild-type resulted in all F1 progeny having the green plants. The mode
of inheritance of the variegated phenotype was determined by self-pollination of the variegated mutant, and by reciprocal crosses between the mutant and wild-type plants. Cross-pollination be-tween the mutant line and wild-type plants was achieved with emasculated flowers, which were covered with paper bags to prevent contamination with foreign pollen. F1 seeds were harvested and
planted as described above. One month after sow-ing, the number of white and green seedlings was recorded. F1-green plants were grown to maturity
in the greenhouse to obtain F2 seeds.
2.4. Microscopic obser6ation
Pieces of leaves were fixed in 2% glutaraldehyde dissolved in 20 mM sodium cacodylate buffer (pH 7.2) for 16 h at 4°C, dehydrated though an ethanol series, and then embedded in Technovit 7100 resin (Kulzer and Co., GmbH, Wehrheim, Germany) as described previously [26]. The samples were sec-tioned (0.6 mm thick) with a glass knife on a
Sorvall MT-2B Ultra Microtome (DuPont, Dela-ware, USA) and dried on cover slips. They were stained with 100 mM/ml DiOC6 (3,3%
2.5. Electron microscopy
The samples were fixed in 3% glutaraldehyde, which was buffered with 20 mM sodium cacody-late at pH 7.0 for 6 h at 4°C, and washed with the same buffer for 16 h at 4°C. Then they were post-fixed with 2% osmium tetroxide in 20 mM cacodylate buffer (pH 7.0) for 6 h at 4°C. The fixed samples were run through an alcohol series and embedded in Spurr’s resin. Ultrathin sections were cut with a diamond knife on an ULTRA-CUT UCT ultramicrotome (Leica, Wien, Austria), transferred to Formvar-coated grids. They were double-stained with 1% uranyl acetate for 15 min at 37°C and with lead citrate solution for 10 min at room temperature. After washing with distilled water, the samples were observed with a JEM-2000 FX II electron microscope (Jeol, Tokyo, Japan).
2.6. RNA gel blot analysis
Young leaves were frozen in liquid nitrogen and total RNA was prepared according to the method of Sambrook et al. [27] with minor modifications. Total RNA (3 mg) was electrophoresed on a
formaldehyde denaturing 1.5% agarose gel in 1×
MOPS buffer (20 mM MOPS-KOH, pH 7.0, 5 mM sodium acetate, and 1 mM EDTA) and then blotted onto a GeneScreen Plus membrane (Du-Pont), using standard protocols [27].
The clones of rbcL which encoded the large subunit of ribulose-1,5-bisphosphate carboxylase; and 16S rDNA which encoded the plastid 16S rRNA were gift of M. Sugiura and M. Sugita in Nagoya Univ. Tobacco psbA which encoded the D1 subunit of the photosystem II reaction center fragments of tobacco (GeneBank Accession No. S54304), cab which encoded a major chlorophyll a/b binding protein Cab21 of tobacco (GeneBank Accession No. X52743), rbcS which encoded the small subunit of ribulose-1,5-bisphosphate car-boxylase of tobacco (GeneBank Accession No. M36685) and cytoplasmic 25S rRNA of tobacco (GeneBank Accession No. X76056) were amplified by polymerase chain reaction (PCR) using Expand High Fidelity PCR system (Boehringer Mannheim) from purified tobacco chloroplast DNA and nuclear genomic DNA using two sets of primer, 5%
-TTATCCATTTGTAGATGGAGCTT-CGATC-3% and 5%
-ATGACTGCAATTTTAGA-GAGACGCGAA-3% for psbA, and 5%
-ATG-GTACGGCCCAGACCGTGTTAAGTAC-3% and 5%
-TCACTTTCCGGGGACAAAGTTTGTGGC-G-3% for cab, and 5% -ATGGCTTCCTCAGTT-CTTTCCTCTGCAG-3% and 5%
-TTAGTAGC-CTTCTGGCTTGTAGGCAATGAA-3%for rbcS,
and 5%-GAATTCACCAAGTGTTGGATTGT-3%
and 5%-ACGAATCGGAGCGACAAAGGG-3%
for 25S rRNA. The fragment was cloned into pCR2.1 vector (Invitrogen) and sequenced by DNA sequencer (model PRISM310, ABI) with Dye Terminator Cycle Sequencing Kit (ABI). The clones were labeled with 32P (6000 Ci/mM) by
Ready To Go-DNA labeling beads (Amersham) and used as hybridization probes.
After hybridization for 24 h at 68°C, the filters were washed in 1×SSPE [0.15 M NaCl, 0.015 M sodium citrate and 0.1% (w/v) SDS] twice at room temperature and three times at 68°C, and exposed to X-ray films for autoradiography. The radioac-tivity of the bands was measured using the BAS 2000 system (Fuji Film Co. Ltd., Tokyo, Japan). Experiments were repeated three times.
3. Results
3.1. Phenotype of the mutant
White areas on the cotyledons initially identified a mutant grown in vitro. As the plant grew, the mutant produced the pale-green and partially light-green leaves with white areas on their edges. The size, shape, and intensity of the green areas of the leaf varied (Fig. 1). The stem and calyces of the mutant have white areas and the stigmata are light yellow, whereas these parts are green in the wild type plant (Fig. 1C and D). The vegetative and reproductive growth of the variegated plant were normal and it produced seeds. The amount of chlorophyll was drastically reduced in the white regions of the mutant (3.691 mg/g fresh weight).
In the mutants, the light green regions found in parts of the pale-green leaves had less chlorophyll (1776941 mg/g fresh weight) than the green
re-gions (2241954 mg/g fresh weight), which had
almost the same amount of chlorophyll as wild type leaves (2334985 mg/g fresh weight).
3.2. Genetic analysis of the mutant
crossed with the wild type, and the leaf color of the progeny was analyzed to determine the genetic basis for this mutant’s variegation. The results of these crosses are presented in Table 1. The F1 generation of the self-pollinated mutant
produced 1169 white (99.5%) and six green (0.5%) seedlings. No segregation for leaf variegation was observed in the F1 and F2
generation. When either the mutant or wild-type plants were used as pollen donors, all the F1 and
F2 progeny were green. On the other hand the
F2 generation progeny of the self-pollinated
F1-green plants of the variegated mutant,
segregated as green and white seedlings.
3.3. Structure of the mutant lea6es
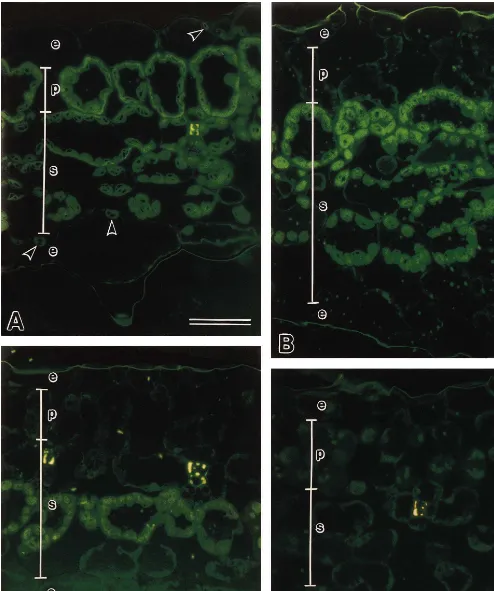
Histological changes in the white areas and adjacent green tissue of variegated leaves were examined microscopically. The fluorochrome DiOC6 stains membranous organelles such as well-developed chloroplasts [26]. In transverse sec-tions of wild-type leaves, all the cells in the pal-isade layer were similar and contained well-developed chloroplasts (Fig. 2A). In the mu-tant leaves, however, the cells in the green (Fig. 2B) and light green (Fig. 2C) regions formed dis-crete sectors in the palisade layer, which contained undeveloped chloroplasts in one or two layers of
Fig. 2. Fluorescence micrographs of leaves from wild-type and variegated plants (N.tabacumL. cv. BY-4) by staining with DiOC6.
Table 1
Number of green and white seedlings obtained from the progeny of the variegated tobacco (Nicotiana tabacum L. cv. BY-4)a
Cross combination Number of flowers Number of plants
Total Green White
Male Variegated
Female
F1generation
18 1169 6 1163 0
V V
6 696 696
WT 0
V 0
V
WT 6 1420 1420 0 0
F2generation of green plants
8 2153 1880
VG-1 273
VG-1 0
VG-2
VG-2 4 186 148 38 0
7 150
VG-3 VG-3 120 30 0
2 151 119
VG-4 32
VG-4 0
8 2321 2321 0 0
VG-1 WT
8 2257 2257 0 0
VG-1 WT
aV, the variegated mutant; WT, wild-type plant; VG, F
2plants derived from self-pollinated F1-green plants of the variegated
mutant.
the spongy parenchyma, and the leaf thickness was irregular. The mutant white regions com-pletely lacked developed chloroplasts (Fig. 2D). In addition, the palisade cells in both the white and green areas of the mutant failed to expand as fully as in the wild type and failed to develop chloro-plasts (Fig. 2B – D). The wild type epidermis had a few well-developed chloroplasts (Fig. 2A), but there were no well-developed chloroplasts in the epidermis of any variegated region of the plant (Fig. 2B – D).
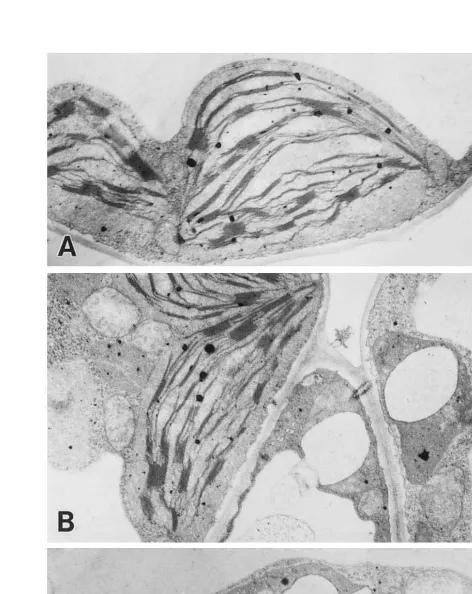
Transmission electron microscopy (TEM) re-vealed that each cell in green regions of the varie-gated plants contained both ordinary chloroplasts and abnormal plastids which had thylakoid mem-branes without grana structures (Fig. 3A and B). On the other hand, white regions of the variegated plants contained only abnormal plastids with ir-regular shapes due to swelling membrane and some electron-dense materials (Fig. 3C).
3.4. Le6els of photosynthetic genes
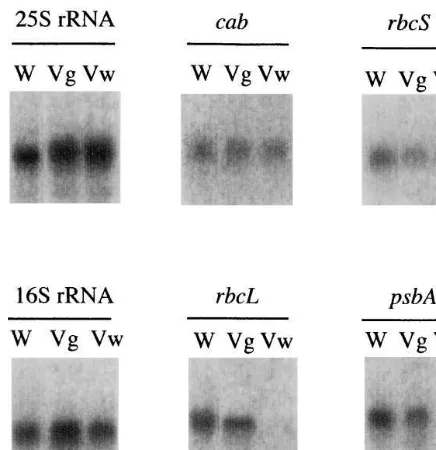
Northern blot analysis was performed with total RNA from the white and green regions of leaves (Fig. 4). Transcript levels ofrbcLand psbA, which are encoded on the chloroplast genome, were abundant in the green regions and drastically re-duced in the white regions. In white regions, the level of plastid-encoded 16S rRNA transcripts was about 70% of the level in green regions. Then, the same amount of transcripts for the
Fig. 4. RNA gel blot analysis of the variegated mutant (N.
tabacumL. cv. BY-4). Total RNA was isolated from leaves of wild-type and variegated seedlings. Total RNA (3 mg) was
separated by electrophoresis, blotted onto a nylon membrane, and hybridized withcab,rbcS,rbcL,psbA, 16S rDNA, or 25S rDNA probes. Transcripts ofrbcLandpsbA mRNA, which are encoded by the chloroplast genome, were drastically reduced in the white region. W, wild-type plants; Vg, green area of the variegated mutant; Vw, white area of the varie-gated mutant.
while the W-W-G mutant produced two green seedlings out of 6000 white seedlings. This analysis indicated that the L2 layer should not be an only source for male and female gametes [12]. Actually invasion of some L3 cells into the L2 layer devel-oping to offspring with the genetic composition of L3 cells was observed in Nicotiana periclinal chimeras [11]. Thus the origin of six green seedlings in the self-cross of F1 generation, might
be deduced as the L3 cells due to the layer inva-sion. Four of the six green plants could set seeds which segregated green and white seedlings in the progeny, however the segregation ratios splitted 7:1 and 4:1 ( green: white). F2 generation progeny
of the self-crossing for S201 also show the similar segregation of green and white [25]. It was sug-gested that the green tissues should be het-erozygous depending on the same mutation gene or genes as the albino mutants, because all be-longed to the family of the S201 line. When either the mutant or wild-type plants were used as pollen donors, all the F1 and F2 progeny were green
indicating of non-maternal inheritance in the var-iegated mutants in reciprocal crosses.
Since the mode of segregation of the periclinal chimera was very similar to the pattern just de-scribed for G-W-G and W-W-G leaves, we exam-ined the anatomy of leaves of the variegated mutant using DiOC6 staining, which stains
well-developed membranes including those of chloro-plasts [26]. No developed chloroplasts were detected in the epidermis layers (L1) or sub-epider-mis tissue (L2) of leaves of the variegated mutant. In addition, one or two layers of spongy par-enchyma (L3) cells from the green sector con-tained undeveloped chloroplasts. This suggests that the variegated mutant has a W-W-G histo-genic composition.
The palisade cells in both the white and the green regions of the variegated mutant failed to expand fully. Several other mutations that block chloroplast development and also affect palisade formation in dicots have been reported in tobacco [12], tomato [28], Arabidopsis [29], and Antir
-rhinum [30]. The gene expression of the variegated mutation may occur very early in chloroplast de-velopment, since no stacked thylakoid membranes were detected and the plastids remained at the proplastid-like stage (Fig. 3B and C). The pale and light green mutant leaves can be attributed to the coded chloroplastic genescabandrbcSwere found
in both green and white regions (Fig. 4).
4. Discussion
anatomical changes in these regions, which have one or two layers of spongy parenchyma cells containing undeveloped chloroplasts, instead of three cell layers with well-developed chloroplasts (Fig. 2B and C). The anatomical changes in the variegated mutant resemble the reduced chloro-phyll content seen in light green areas of other variegation mutations [7,13].
The levels of rbcS and cab transcription in the variegated mutant were different from that of the
immutants, a variegated Arabidopsis [21]. For ex-ample, those levels of theimmutantsbecame lower in the white tissues, while equal amounts of these RNAs were observed in the white sectors of varie-gated tobacco mutant. Transcription levels of
rbcL and psbA in white tissues of the immutants
were slightly lower than the wild type, however those of the tobacco mutant showed drastic de-crease at the white tissues (Fig. 4). This pattern of chloroplast gene expression has not yet been re-ported in variegated mutation, but it resembles that of the rpoB deletion transformant [31]. The
rpoBgene is plastid-localized and nuclear-encoded RNA polymerase. The deletion of rpoB plants contained a uniformly transgenic plastid genome and resulted in a pigment-deficient phenotype. Our data indicate that variegated mutation may occur in the gene or genes related to chloroplast mRNA regulation specifically. Regulation system on chloroplast encoded genes includes transcrip-tional regulations, post-transcriptranscrip-tional regulations and DNA copy number regulations. Future analyses for the variegated mutant will clarify these detail mechanisms between the variegation and chloroplastic gene regulations.
Acknowledgements
We are grateful to Dr Toshiaki Kameya of Tohoku University for helpful comments on this manuscript. We also thank Dr Masahiro Sugiura and Dr Mamoru Sugita of Nagoya University for the gift of the tobacco chloroplast DNA. This work was supported by a grant-in-aid of Scientific Research (No. 09760010 and 11151230) from the Ministry of Education, Science, and Culture, of Japan, a grant for genome research from RIKEN, and a Science and Technology Agency fellowship from the Japan International Science and Tech-nology Exchange Center.
References
[1] T. Bo¨rner, B.B. Sears, Plastome mutants, Plant Mol. Biol. Rep. 4 (1986) 69 – 92.
[2] A. Barkan, Nuclear mutants of maize with defects in chloroplast polysome assembly have altered chloroplast RNA metabolism, Plant Cell 5 (1993) 389 – 402. [3] D.L. Rousell, D.L. Thompson, S.G. Pallardy, D. Miles,
K.J. Newton, Chloroplast structure and function is al-tered in the NCS2 maize mitochondrial mutant, Plant Physiol. 96 (1991) 232 – 238.
[4] S.G. Wildman, C. Lu-Liao, F. Wong-Staal, Maternal inheritance, cytology of defective chloroplasts in a varie-gated mutant of Nicotiana tabacum, Planta 113 (1973) 293 – 312.
[5] F. Wong-Staal, S.G. Wildman, Identification of a muta-tion in chloroplast DNA correlated with formamuta-tion of defective chloroplasts in a variegated mutant of Nico
-tiana tabacum, Planta 113 (1973) 313 – 326.
[6] A.B. Bonnema, N. Castillo, N. Reiter, M. Cunningham, H.P. Adams, M. O’Connell, Molecular and ultrastruc-tural analysis of a nonchromosomal variegated mutant: tomato mitochondrial mutants that cause abnormal leaf development, Plant Physiol. 109 (1995) 385 – 392. [7] H.T. Bonnett, I. Djurberg, M. Fajardo, K. Glimelius, A
mutation causing variegation and abnormal development in tobacco is associated with an altered mitochondrial DNA, Plant J. 3 (1993) 519 – 525.
[8] K.J. Newton, E.H. Coe Jr, Mitochondrial changes in abnormal growth (nonchromosomal stripe) mutants in maize, Proc. Natl. Acad. Sci. USA 83 (1986) 7363 – 7366. [9] W. Sakamoto, H. Kondo, M. Murata, F. Motoyoshi, Altered mitochondrial gene expression in a maternal distorted leaf mutant ofArabidopsis induced by chloro-plast mutator, Plant Cell 8 (1996) 1377 – 1390.
[10] D. Wu, D.A. Wright, C. Wetzel, D.F. Voytas, S. Roder-mel, The immutants variegation locus of Arabidopsis
defines a mitochondrial alternative oxidase homolog that functions during early chloroplast biogenesis, Plant Cell 11 (1999) 43 – 55.
[11] M. Margotrigiano, R. Bernatzky, Arrangement of cell layers in the shoot apical meristems of periclinal chimeras influences cell, Plant J. 7 (1995) 193 – 202. [12] R.N. Stewart, L.G. Burk, Independence of tissues
derived from apical layers in ontogeny of the tobacco leaf and ovary, Am. J. Bot. 58 (1970) 1010 – 1016. [13] S. Poethig, Genetic mosaics and cell lineage in plants,
Trends Genet. 5 (1989) 273 – 277.
[14] R.N. Stewart, The origin and transmission of a series of plastogene mutants inDianthusandEuphorbia, Genetics 52 (1965) 925 – 947.
[15] C. Aoki, T. Wada, T. Nishimura, K. Hattori, Character-ization and inheritance of variegated-leaf mutant in
Petunia hybrida, Breed. Sci. 45 (1995) 31 – 35.
[16] J. Gu, D. Miles, K.J. Newton, Analysis of leaf sectors in the NCS6 mitochondrial mutant of maize, Plant Cell 5 (1993) 963 – 971.
[18] J.M. Martinez-Zapater, P. Gil, J. Capel, C.R. Somerville, Mutations at the arabidopsis CHM locus promote rear-rangements of the mitochondrial genome, Plant Cell 4 (1992) 889 – 899.
[19] D. McCormac, J.J. Boinski, V.C. Ramsperger, J.O. Berry, C4 gene expression in photosynthetic and non-photosynthetic leaf regions ofAmaranthus tricolor, Plant Physiol. 114 (1997) 801 – 815.
[20] W.C. Taylor, Regulatory interactions between nuclear and plastid genomes, Ann. Rev. Plant Mol. Biol. 40 (1989) 211 – 233.
[21] C.M. Wetzel, C.Z. Jiang, L.J. Meehan, D.F. Voytas, S.R. Rodermel, Nuclear-organelle interactions: the im
-mutantsvariegation mutant of Arabidopsisis plastid au-tonomous and impaired in carotenoid biosynthesis, Plant J. 6 (1994) 161 – 175.
[22] T. Abe, S. Yoshida, T. Sakamoto, T. Kameya, S. Ki-tayama, N. Inabe, M. Kase, A. Goto, Y. Yano, Is irradiation of heavy ion beams at specific stages of the fertilization cycle of plants effective for mutagenesis?, in: K. Oono, F. Takaiwa (Eds.), Modification of Gene Expression and Non-Mendelian Inheritance, NIAR, Japan, 1995, pp. 469 – 477.
[23] T. Murashige, F. Skoog, A revised medium for rapid growth and bioassay with tobacco tissue cultures, Phys-iol. Plant 15 (1962) 473 – 477.
[24] D.I. Aron, Copper enzymes in isolated chloroplasts: polyphenoloxidase in Beta 6ulgaris, Plant Physiol. 24
(1949) 1 – 15.
[25] C.H. Bae, T. Abe, T. Matsuyama, T. Nakano, S. Yoshida, Effect of heavy-ion beam irradiation on muta-tion inducmuta-tion in plants at pollinamuta-tion stage: Selecmuta-tion of salt-tolerant tobacco, Breed. Sci. 47 (Suppl. 1) (1997) S218.
[26] M. Fujie, H. Kuroiwa, S. Kawano, S. Mutoh, T. Kuroiwa, Behavior of organelles and their nucleoids in the shoot apical meristem during leaf development in
Arabidopsis thalianaL, Planta 194 (1994) 395 – 405. [27] J. Sambrook, E.F. Fritsch, T. Maniatis, Analysis of
RNA, in: J. Sambrook, E.F. Fritsh, T. Maniatis (Eds.), Molecular Cloning, A Laboratory Manual, second ed., Cold Spring Harbor Laboratory Press, NY, 1989, pp. 7.37 – 7.52.
[28] J.S. Keddie, B. Carroll, J.D.C. Jones, W. Gruissem, The DCL gene of tomato is required for chloroplast develop-ment and palisade cell morphogenesis in leaves, EMBO J. 15 (1996) 4208 – 4217.
[29] R.S. Reiter, S.A. Coomber, T.M. Bourett, G.E. Bartley, P.A. Scolnik, Control of leaf and chloroplast develop-ment by the Arabidopsis gene pale cress, Plant Cell 6 (1994) 1253 – 1264.
[30] M. Chatterjee, S. Sparvoli, C. Edmunds, P. Garosi, K. Findlay, C. Martin, DAG, a gene required for chloro-plast differentiation and palisade development in Antir
-rhinum majus, EMBO J. 15 (1996) 4194 – 4207.
[31] L.A. Allison, L.D. Simon, P. Maliga, Deletion of rpoB
reveals a second distinct transcription system in plastids of higher plants, EMBO J. 15 (1996) 2802 – 2809.