Pepper gene encoding a basic
b
-1,3-glucanase is differentially
expressed in pepper tissues upon pathogen infection and ethephon
or methyl jasmonate treatment
Ho Won Jung, Byung Kook Hwang *
Molecular Plant Pathology Laboratory,Department of Agricultural Biology,Korea Uni6ersity,Seoul136-701,South Korea
Received 21 February 2000; received in revised form 17 May 2000; accepted 3 July 2000
Abstract
A basicb-1,3-glucanase cDNA clone (CABGLU) was isolated from the cDNA library constructed from hypersensitive response lesions of pepper leaves infected with avirulent strain of Xanthomonas campestris pv. 6esicatoria. The deduced polypeptide of
CABGLU which contains a C-terminal extension N-glycosylated at a single site characterized as typical structure of class I
b-1,3-glucanase has a high level of identity with tobacco basicb-1,3-glucanase (77.4%), but only a moderate level of identity with tomato acidic b-1,3-glucanase (42.6%). Genomic DNA gel blot analysis indicates that the pepper genome contains one or two
b-1,3-glucanase copy genes. Transcripts of theCABGLUgene were more induced in incompatible interactions than in compatible interactions, when inoculated withX.campestrispv.6esicatoriaorPhytophthora capsici. Accumulation ofCABGLUmRNA was strongly induced in pepper leaves by both ethephon and methyl jasmonate. TheCABGLU mRNA was constitutively expressed only in the roots of all the plant organs. These data indicate that the basic b-1,3-glucanase gene may be induced by pathogen attack and abiotic stresses. © 2000 Elsevier Science Ireland Ltd. All rights reserved.
Keywords:Basicb-1,3-glucanase;Capsicum annuum; Ethephon; Methyl jasmonate;Phytophthora capsici;Xanthomonas campestrispv.6esicatoria
www.elsevier.com/locate/plantsci
1. Introduction
Plants respond to pathogen attack or external environmental stresses by producing a diverse set of proteins. These proteins, which have been most extensively studied in tobacco plants infected with tobacco mosaic virus (TMV), are commonly re-ferred to as pathogenesis-related (PR) proteins [1]. In particular, chitinase and b-1,3-glucanase have long been suggested to be associated with the antifungal defenses of plants [2,3].b-1,3-Glucanase
hydrolyses the b-1,3-linked glucans, major compo-nents of cell wall of oomycetes and synergistically acts with chitinase to inhibit fungal growth in vitro [3,4]. The enzyme may also mediate in plant defense by releasing glucan fragments from fungal or plant cell walls as signal molecules that can activate a variety of plant defenses [5,6]. In addi-tion to pathogen attack, the expression of b-1, 3-glucanases has been shown to be induced by abiotic elicitors such as ethylene [6], salicylic acid [7], and methyl jasmonate [8]. Additionally, b -1,3-glucanases have been involved in several physio-logical and developmental processes such as microsporogenesis [9], seed germination [10], pol-len development [11], and fruit development [12].
b-1,3-Glucanase has been demonstrated to exist in multiple isoforms in a number of plant species
Nucleotide and amino acid sequence data has been deposited in EMBL/GenBank database under accession number AF227953.
* Corresponding author. Tel.: +82-2-32903061; fax: + 82-2-9251970.
E-mail address:[email protected] (B.K. Hwang).
including tobacco [13], potato [14], bean [15], and rice [16]. In our earlier study, two acidicb -1,3-glu-canase isoforms and six basic b-1,3-glucanase iso-forms were detected in pepper stems infected with Phytophthora capsici [17]. We further isolated a b-1,3-glucanase with antifungal activity from pep-per stems [4]. The two acidic and six basic b -1,3-glucanases were also induced and accumulated in pepper leaves infected with Xanthomonas campes -trispv.6esicatoria[18]. However, the role ofb -1,3-glucanase either in defense responses or in physiological and developmental processes has not been well understood, since the enzyme activity exists in several isoforms that differ in amino acid sequences, cellular localization and inducibility [19].
Based on amino acid sequence identities, b -1,3-glucanases from tobacco have been classified into three structural classes [20]. The class I b -1,3-glucanase is approximately 33 kDa, basic proteins, and accumulates intracellularly at a site presumed to be vacuoles [21]. This vacuolar localization correlates with the presence of a distinctive C-ter-minal extension that contains sorting information for proper targeting to the vacuole [19]. Class II b-1,3-glucanase includes the acidic isoforms such as PR-2a, PR-2b, and PR-2c, formerly named PR-2, PR-N, and PR-O, respectively [13,20]. The single representative of the class III isoforms is PR-Q’ found in TMV-infected tobacco leaves [20]. Both class II and III b-1,3-glucanases accumulate predominantly in the extracellular space, i.e. the apoplast. In some cases, the acidic b -1,3-glu-canases were shown to be present in intercellular washing fluid (IWF) from pepper tissues infected with pathogens [17,18].
To gain a better understanding of the role of PR-proteins including b-1,3-glucanase, we con-structed a cDNA library from pepper leaves infected with avirulent strain Bv5-4a ofX.campes -tris pv. 6esicatoria [22]. More recently, some
PR-gene clones encoding b-1,3-glucanase, chiti-nase, thionin, and lipid transfer protein were isolated from this cDNA library using the differ-ential hybridization technique [23]. In this paper, we report on the isolation and sequence analysis of a full-length cDNA clone encoding a basic b-1, 3-glucanase from pepper leaves. We further show that expression of the basic b-1,3-glucanase gene is induced in pepper leaves and stems by pathogen infection and abiotic elicitor treatments.
2. Materials and methods
2.1. Plant and pathogens
Pepper (Capsicum annuum L., cv. Hanbyul) plants were raised in a growth room to the four or six-leaf stages, as previously described [24].
The two strains Ds1 and Bv5-4a of X. campes -trispv.6esicatoria that were virulent and avirulent
to the pepper cultivar Hanbyul, respectively, were used in this study [25]. The bacteria strains were grown overnight in YN medium (5 g yeast extract, 8 g nutrient broth and 1 l H2O) at 28°C. The bacteria were harvested from broth cultures by centrifugation, and suspended at 108 CFU/ml in sterile tap water prior to inoculation. Inoculation with X. campestris pv. 6esicatoria was done by
vacuum-infiltrating the bacterial suspension into the abaxial side of fully expanded leaves of pepper plants at six-leaf stage with an automizer con-nected to a compressor.
Two isolates, S197 and CBS 178.26, ofPhytoph -thora capsici which are virulent and avirulent to pepper cultivar Hanbyul, respectively, were used in this study [26]. The fungal pathogens were grown on oatmeal plates for 10 days at 28°C in dark. Zoospores were harvested from sporulating plates, and adjusted to 105 zoospores per ml with sterile tap water prior to inoculation. A small quantity of cotton soaked in zoospore suspensions was placed on the bottom region of each pepper stem. The inoculated sites were then covered with plastic tape to maintain moist conditions. Plant samples were taken at various time intervals after inoculation. Harvested leaves and stems were im-mediately frozen in liquid nitrogen, and stored at
−70°C until isolated for total RNA.
2.2. Abiotic elicitor treatment
molarity to ethephon to evaluate the effect of them. The pepper plants were incubated in a growth-room at 28°C with 16-h day length. Plants applied with ethephon or MeJA were enclosed in a vinyl bag. Pepper leaf samples were harvested at various time intervals after application, immedi-ately frozen in liquid nitrogen, and stored at −
70°C until isolated for total RNA.
2.3. Isolation of a basic b-1,3-glucanase cDNA clone (CABGLU)
To isolate pathogenesis related genes from pep-per leaves, individual cDNA clones strongly or differentially expressed in the infected leaves were selected using a differential hybridization tech-nique [23]. These cDNA genes were designated CAIs (Capsicum annuum induced genes). After the 5%partial sequencing ofCAIgenes using automatic DNA sequencer (ABI310, Applied Biosystem), the 5% end partial nucleotide and deduced peptide se-quences obtained were analyzed using the PC/
Gene software system and BLAST network services at the National Center for Biotechnology Information (NCBI) [27]. The partial nucleotide sequence data of the CAI20 have been deposited in the EMBL/GenBank database under accession number AF082724 [23].
2.4. DNA sequencing and analysis
To determine a full-length sequence of CAI20, the cDNA in the pBluescript SK(−) was se-quenced on ABI310 DNA sequencer (Applied Biosystem) using Thermo-cycle sequenase kit (Amersham) with T3 or T7 primer according to manufacture’s instruction. Progressive deletions were obtained by using Erase-a-Base system (Promega, Madison, WI, USA). The amino acid alignments were manually adjusted to compare cDNA clones with those of other organisms.
2.5. RNA gel blot analysis
Total RNA was extracted from pepper leaves, stems, root, flowers, and fruits by the guanidinium thiocyanate method [28]. The concentration and integrity of total RNA in individual extracts were determined by UV absorbance and staining with ethidium bromide, respectively.
Total RNA (30mg) was denatured by heating at 65°C for 10 min in a formaldehyde gel-loading buffer, separated by electrophoresis on 7.4% form-aldehyde gels, and transferred to nylon mem-branes (Hybond N+, Amersham) [29]. RNA was
cross-linked on the blots by UV illumination. An EcoRI/XhoI restriction fragment in pBluescript SK(−) recombinant plasmid carrying the pepper b-1,3-glucanase gene was 32P-labeled with a ran-dom prime kit (Boehringer Mannheim). Prehy-bridization and hyPrehy-bridization was performed at 65°C in 5% (w/v) dextran sulfate, 0.25 M disodium phosphate pH 7.2, 7% (w/v) sodium dodecyl sul-fate (SDS), and 1 mM EDTA. The membranes were washed twice with 2X SSC and 0.1% SDS for 10 min at room temperature and finally several times with 0.1×SSC and 0.1% SDS for 5 min at 65°C. To control equal transfer of RNA, the blots were co-hybridized with a C. annuum 25S rRNA probe.
2.6. DNA gel blot analysis
C. annuum genomic DNA was prepared from
young leaves, as previously described by Hong et al. [30]. Each gram of tissue ground under liquid nitrogen was suspended in 3 ml extraction buffer [8.0 M urea, 50 mM Tris – HCl (pH 7.5), 20 mM EATA, 350 mM NaCl, 2% (w/v) SDS, 5% (v/v) phenol and 20 mM EDTA 2-mercaptoethanol]. After successive extractions with phenol/ chloro-form/isoamylalcohol(25:24:1, v/v/v), the high molecular weight genomic DNA was recovered by spooling. Twenty micrograms of genomic DNA were digested with appropriate restriction en-zymes, according to the protocols described by Sambrook et al. [29]. After ethanol precipitation, completely digested genomic DNA was resus-pended in 10 mM Tris and 1 mM EDTA prior to gel electrophoresis. The DNA fragments were sep-arated on 0.8% agarose gel. The DNA was trans-ferred to Hybond N+ (Amersham) membrane, and hybridized to 32P-labeled CABGLU gene probe at 65°C as described above.
3. Results
3.1. Sequence analysis of CABGLU cDNA
encoding a putative basicb-1,3-glucanase that was expressed in pepper leaves undergoing HR. To determine a full-length nucleotide sequence of the cDNA clone, one set of nested deletions was constructed. The full-length cDNA sequence was designated as CABGLU (C. annuum basic b-1,3-glucanase). The CABGLU gene contains 1332 bp with an apparent single open reading frame initiated by ATG codon 25 base pairs far from the 5%end (data not shown). The open reading
frame ends with two successive stop codons at position 1104 (TGA and TAA). There is a potential polyadenylation signal AATAAA starting 42 bases upstream from this polyadenylation tail. The
predicted CABGLU protein has a typical
hydrophobic signal peptide cleavage site located between residues 20 (glycine) and 21 (glutamine). The pepper CABGLU cDNA encodes a putative polypeptide of 359 amino acids with a predicted molecular mass of 39 226 Da and a predicted isoelectric point (pI) of 9.46. Moreover, the
CABGLU gene has a C-terminal extension signal
sequence, which is necessary for targeting to the vacuole [9,31]. An interesting feature in the amino acid sequence of theCABGLUgene is the existence of a consensus sequence for N-glycosylation (Asn-Ala-Thr) at amino acid positions 349 – 351. The predicted polypeptide sequence of the
CABGLUcDNA encoding a putative pepper basic
b-1,3-glucanase was compared with the previously sequenced b-1,3-glucanases from other plant species (Fig. 1). The amino acid sequence homology search revealed that the protein encoded
by CABGLU shares a significant identity with
other known plant basic b-1,3-glucanases. The basic b-1,3-glucanase of pepper has 77.4% identity with a tobacco (Nicotiana plumbaginifolia) basic b-1,3-glucanase [32,33], 64.6% identity with a potato (Solanum tuberosom) basic b-1,3-glucanase
[34], and 58.7% identity with a tomato
(Lycopersicon esculentum) basic b-1,3-glucanase [35]. The amino acid sequence of CABGLU has only 42.6% identity with tomato tomQ’a gene encoding acidic b-1,3-glucanase [19]. The alignment shows that the basicb-1,3-glucanase has a C-terminal extension which does not contain acidic b-1,3-glucanase.
3.2. DNA gel blot analysis of CABGLU cDNA
DNA gel blot analysis was performed on C.
annuum genomic DNA digested with EcoRI,
EcoRV and XbaI which do not possess specific recognition sites in theCABGLUcDNA sequence. To avoid cross-hybridization of several isoforms of pepper b-1,3-glucanase gene, hybridization and blot washing conditions were performed at high stringency. Two major bands were detected in EcoRI digests, while one major band was detected in both EcoRV and XbaI digests (Fig. 2). These results indicated that at high stringency, the CABGLU gene is present as one or two copies per
C. annuum genome.
3.3. Organ-specific expression of CABGLU gene
To examine organ-specific expression of the
CABGLU gene, RNA gel blot analysis was
per-formed using transcripts of various tissues of pep-per plants (Fig. 3). The CABGLU mRNAs were not detectable in healthy leaves and fruits. In contrast, the expression of the CABGLUgene was strong in roots, but remained at low levels in stems and flowers.
3.4. Expression of CABGLU gene by pathogen
infection
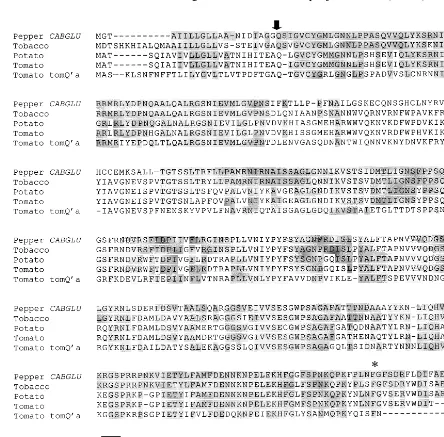
To studyCABGLU gene expression after bacte-rial infection, pepper leaves were inoculated with the strains Ds1 or Bv5-4a of X. campestris pv.
6esicatoria which were virulent or avirulent to pepper cultivar Hanbyul, respectively. Fig. 4 shows the CABGLU mRNA expression pattern in the compatible and incompatible interactions be-tween pepper andX.campestrispv.6esicatoria. No transcripts homologous to CABGLU gene were detected in the healthy pepper leaves. In the com-patible interaction, accumulation of CABGLU mRNA was slightly detected at 18 h after inocula-tion, continued to rise thereafter, and remained at a high level to 30 h after inoculation. In the incompatible interaction, accumulation of
CABGLU mRNA was more inducible than in the
compatible interaction. In particular, the accumu-lation of CABGLUmRNA drastically increased in the infected leaves at 24 h after inoculation of the avirulent strain.
virulent or avirulent to pepper cultivar Hanbyul, respectively. CABGLU mRNA was slightly de-tected in healthy or wounded pepper stems. In the compatible and incompatible interactions, some induction of CABGLU mRNA was found at 24 h after inoculation. The highest level of the tran-scripts was detected in the compatible response at 48 h after inoculation, but the mRNA levels grad-ually declined from 48 to 96 h after inoculation, when the infected stems completely withered. In contrast, the mRNA levels of CABGLU gene in
the incompatible response gradually increased from 24 to 72 h after inoculation.
3.5.Induction of CABGLU mRNA by ethephon and
methyl jasmonate
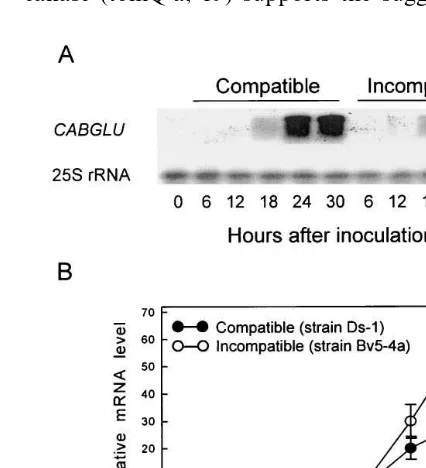
The levels of expression of the CABGLU mRNA in the pepper leaves by abiotic elicitor treatment were examined by RNA gel blot analy-sis (Fig. 6). Following treatment with ethephon and methyl jasmonate (MeJA), the CABGLU
Fig. 2. Genomic DNA gel blot analysis of CABGLU gene encoding a basic b-1,3-glucanase in pepper plants. The en-zymes that were used for digestion of genomic DNA are indicated above each lane. The numbers on the left indicate the length of the fragments in kilobases. Twenty micrograms of digested genomic DNA were separated in each lane.
remained at a high level by 24 h after treatment (Fig. 7A). In pepper leaves treated with MeJA,
CABGLU transcripts were detected at 12 h after
treatment and increased drastically thereafter (Fig. 7B).
4. Discussion
Using a differential hybridization strategy for isolation of pathogenesis-related genes, a basic b-1,3-glucanase gene (CABGLU) was isolated from the cDNA library made from mRNAs of pepper leaves infected with avirulent strain Bv5-4a of X. campestris pv. 6esicatoria. The deduced
amino acid sequence of CABGLUcDNA is highly conserved in the coding region based on the align-ments with sequences of the three basic b -1,3-glu-canases of other plants. The 42.6% identity of the pepper CABGLU with tomato acidic b -1,3-glu-canase (tomQ’a, 19) supports the suggestion that
Fig. 4. RNA gel blot analysis of transcript levels of pepper
CABGLU gene in leaves inoculated with virulent strain Ds-1 and avirulent strain Bv5-4a of Xanthomonas campestris pv. vesicatoria. (A) Northern blot analysis of induction of
CABGLUmRNA at various times after inoculation. (B) The relative induction of CABGLU mRNA levels between com-patible and incomcom-patible interactions, as estimated by densit-ometry of autoradiographic signals obtained from panel A. Thirty micrograms of total RNA from each sample were separated in each lane. ACapsicum annuum25S rRNA probe was used as an internal standard. The Northern blot analysis was repeated three times with similar results.
mRNA was strongly detected in pepper leaves at 24 h after treatment, as shown in Fig. 6. However, after treatment with SA, BABA and BTH, tran-scripts of CABGLU gene did not accumulate in pepper leaves. Hydrochloric acid (HCl) and phos-phonic acid (H3PO3) as breakdown products of ethephon did not induce transcripts of CABGLU gene. A time course analysis was performed to further investigate in detail the effect of ethephon and MeJA in inducing theCABGLU mRNA gene (Fig. 7). The transcripts ofCABGLUgene began to accumulate in pepper leaves at 6 h after treatment with ethephon, drastically increased 6 – 12 h, and
Fig. 3. Northern blot analysis ofCABGLUmRNA expression in various tissues of pepper plants. Thirty micrograms of total RNA from each sample were separated in each lane. An
EcoRI/XhoI fragment of the putative pepper CABGLU
Fig. 5. RNA gel blot analysis of transcript levels of pepper
CABGLUgene in stems inoculated with virulent isolate S197 and avirulent isolate CBS 178.26 ofPhytophthora capsici. (A) Northern blot analysis of induction of CABGLU gene at various times after inoculation. (B) The relative induction of
CABGLU mRNA levels between compatible and incompat-ible interactions, as estimated by densitometry of autoradio-graphic signals obtained from panel A. Total RNA was isolated from pepper stems at various times after inoculation and 24 h after wounding. Thirty micrograms of total RNA from each sample were separated in each lane. A Capsicum annuum 25S rRNA probe was used as an internal standard. The Northern blot analysis was repeated three times with similar results.
Fig. 6. RNA gel blot analysis ofCABGLUmRNA expression in pepper leaves in response to various abiotic stresses. Total RNA was isolated from pepper leaves 24 h after treatment with ethephon (10 mM), salicylic acid (5 mM), methyl jas-monate (100 mM), DL-b-amino-n-butyric acid (2000mg/ml), benzothiadiazole CGA245704 (20 mg/ml), phosphonic acid (H3PO3, 10 mM) and hydrochloric acid (HCl, 10 mM). Thirty
micrograms of total RNA from each sample were separated in each lane. ACapsicum annuum25S rRNA probe was used as an internal standard. The Northern blot analysis was repeated three times with similar results.
Fig. 7. RNA gel blot analysis ofCABGLUmRNA expression in pepper leaves treated with ethephon (A) or methyl jas-monate (B). Total RNA was isolated from pepper leaves at various times after treatment with ethephon (10 mM) or methyl jasmonate (100mM). Thirty micrograms of total RNA from each sample were separated in each lane. A Capsicum annuum 25S rRNA probe was used as an internal standard. The Northern blot analysis was repeated three times with similar results.
tomato b-1,3-glucanase genes, the deduced amino acid sequence ofCABGLU gene has a single puta-tiveN-glycosylation site, Asn-Ala-Thr (N-A-T), at positions 349 – 351, and a glycine (G) adjacent to phenylalanine (F) in the C-terminal extension. The sequence data suggests that processing of the
CABGLU pepper b-1,3-glucanase may lead to the
N-glycosylation of the C-terminal extension. The C-terminal extension N-glycosylated is known to be removed to give the mature protein that is not glycosylated [37]. The CABGLUpepper b -1,3-glu-canase may be processed by the cleavage of an N-terminal signal peptide and a C-terminal exten-sion sequence N-glycosylated, as described for other plant species [36,38]. In our early studies on PR protein accumulation during pathogen attack or abiotic stress [4,17,18], two acidic b -1,3-glu-canase and six basicb-1,3-glucanase were found to be induced and accumulate in pepper leaves or stems infected by pathogens. In particular, purified 34 kDab-1,3-glucanase which is located inside the protoplast and inhibited hyphal growth of the P. capsicidid not show antifungal activity against the chitin-containing fungi such as Alternaria mali,
Colletotrichum gloeosporioides and Magnaporthe
grisea [4].
Northern hybridization was performed with the pepper basic b-1,3-glucanase gene (CABGLU) to examine the expression pattern in pepper leaves infected withX.campestris pv.6esicatoria. At 30 h
after inoculation with virulent strain Ds-1, typical susceptible lesions were not yet observed in pepper leaves. On the other hand, a hypersensitive re-sponse was observed on pepper leaves infected with avirulent strain Bv5-4a at 18 h after
inocula-tion. Accumulation of CABGLU mRNA was
strongly induced in pepper leaves infected with X. campestris pv. 6esicatoria, irrespective of
compat-ible or incompatcompat-ible interaction. However,
accu-mulation of CABGLU mRNA drastically
increased in pepper leaves infected with the aviru-lent bacteria strain, suggesting that b -1,3-glu-canase may participate in plant defense response, as well as infection process. This result is sup-ported by the finding that b-1,3-glucanase was similarly induced by both virulent and avirulent pathogens on plant tissues [3,36]. In pepper stems infected with the oomycetes P. capsici, the accu-mulation ofCABGLUmRNA was greatly reduced in compatible interactions between 48 and 96 h
after inoculation. The decrease of transcripts of CABGLU at late infection stages may result from a severe deterioration of the infected stems. In conclusion, pepper basic b-1,3-glucanase may me-diate a part of the defense responses to pathogen infections.
To determine whether or not transcripts of
CABGLU were expressed by abiotic elicitors, we
also applied ethephon, SA, MeJA, BABA, and BTH on pepper leaves. In this study, ethephon and MeJA activated accumulation of b -1,3-glu-canase mRNA, suggesting that ethylene released from ethephon may activate basic isoforms of PR-proteins [39]. In contrast, exogenous SA, BABA, and BTH did not activate the pepper basic b-1,3-glucanase gene in pepper leaves. In particu-lar, the accumulation of CABGLU mRNAs was greatly induced in pepper leaves treated with either ethephon or MeJA. This result suggests that ethyl-ene and MeJA may be signal molecules to activate the basic b-1,3-glucanase gene in pepper leaves. These observations also indicate that SA indepen-dent pathway may be involved in transcription of the pepper basic b-1,3-glucanase gene.
In general, intracellular basic PR-proteins have been known to be induced in response to develop-mental signals [39]. In this study, accumulation of
CABGLU mRNA was constitutively or strongly
detected in healthy pepper roots, suggesting that b-1,3-glucanase may play a significant role in pro-tection of susceptible tissues against pathogen at-tack in a preemptive fashion [40]. The CABGLU gene was also weakly expressed in flowers of pep-per plants, which may indicate the involvement of the hydrolase in the formation of reproductive organs such as flowers.
References
[1] L.C. Van Loon, Y.A.M. Gerritsen, C.E. Ritter, Identifi-cation, purifiIdentifi-cation, and characterization of pathogene-sis-related proteins from virus-infected Samsun NN tobacco leaves, Plant Mol. Biol. 9 (1987) 593 – 609. [2] F.B. Abeles, R.P. Bosshart, L.E. Forrence, W.H. Habig,
Preparation and purification of glucanase and chitinase from bean leaves, Plant Physiol. 47 (1971) 129 – 134. [3] F. Mauch, B. Mauch-Mani, T. Boller, Antifungal
hydro-lases in pea tissue, Plant Physiol. 88 (1988) 936 – 942. [4] Y.J. Kim, B.K. Hwang, Isolation of a basic
34-kilodal-tonb-1,3-glucanase with inhibitory activity againstPhy
[5] N.T. Keen, M. Yoshikawa, b-1,3-Endoglucanase from soybean releases elicitor-active carbohydrates from fun-gus cell wall, Plant Physiol. 71 (1983) 460 – 465. [6] Y. Takeuchi, M. Yoshikawa, G. Takeba, T. Kunisuke,
D. Shibata, O. Horino, Molecular cloning and ethylene induction of mRNA encoding a phytoalexin elicitor-re-leasing factor, b-1,3-endoglucanase, in soybean, Plant Physiol. 93 (1990) 673 – 682.
[7] E.R. Ward, S.J. Uknes, S.C. Williams, S.S. Dincher, D.L. Wiederhold, D.C. Alexander, P. Ahl-Goy, J.P. Metraux, J. Ryals, Coordinate gene activity in response to agents that induce systemic acquired resistance, Plant Cell 3 (1991) 1085 – 1094.
[8] M. Rickauer, W. Brodschelm, A. Bottin, C. Veronesi, H. Grimal, M.T. Esquerre-Tugaye, The jasmonate pathway is involved differentially in the regulation of different defence response in tobacco cells, Planta 202 (1997) 155 – 162.
[9] D. Worrall, D.L. Hird, R. Hodge, W. Paul, J. Draper, R. Scott, Premature dissolution of the microsporocyte callose wall causes male sterility in transgenic tobacco, Plant Cell 4 (1992) 759 – 771.
[10] G. Leubner-Metzger, C. Frundt, R. Vogeli-Lange, F. Meins, Class Ib-1,3-glucanases in endosperm of tobacco during germination, Plant Physiol. 109 (1995) 751 – 759. [11] P.A. Bucciaglia, A.G. Smith, Cloning and characteriza-tion of Tag 1, a tobacco anther b-1,3-glucanase ex-pressed during tetrad dissolution, Plant Mol. Biol. 24 (1994) 903 – 914.
[12] T.G. McCollum, H. Doostdar, R.T. Mayer, R.E. Mc-Donald, Characterization of chitinase and b -1,3-glu-canases in grape fruit flavedo during fruit development, Physiol. Plant. 99 (1997) 486 – 494.
[13] S. Kauffmann, M. Legrand, P. Geoffroy, B. Fritig, Biological function of ‘pathogenesis-related’ proteins: four PR proteins of tobacco have 1,3-b-glucanase activ-ity, EMBO J. 6 (1987) 3209 – 3212.
[14] E. Kombrink, M. Schroder, K. Hahlbrock, Several ‘pathogenesis-related’ proteins in potato are 1, 3-b -glu-canases and chitinase, Proc. Natl. Acad. Sci. USA 85 (1988) 782 – 786.
[15] A. Awade, M. De Tapia, L. Didierjean, G. Burkard, Biologcal function of bean pathogenesis-related (PR3 and PR4) protein, Plant Sci. 63 (1989) 121 – 130. [16] C.S. Anuratha, K.C. Zen, K.C. Cole, T. Mew, S.
Muthukrishnan, Induction of chitinases and b -1,3-glu-canases inRhizoctonia solani-infected rice plants: isola-tion of an infecisola-tion-related chitinase cDNA clone, Physiol. Plant. 97 (1996) 39 – 46.
[17] Y.J. Kim, B.K. Hwang, Differential accumulation of
b-1,3-glucanase and chitinase isoforms in pepper stems infected by compatible and incompatible isolates ofPhy
-tophthora capsici, Physiol. Mol. Plant Pathol. 45 (1994) 195 – 209.
[18] Y.K. Lee, B.K. Hwang, Differential induction and accu-mulation of b-1,3-glucanase and chitinase isoforms in the intercellular space and leaf tissues of pepper by
Xanthomonas campestrispv.6esicatoriainfection, J. Phy-topathol. 144 (1996) 79 – 87.
[19] C. Domingo, V. Conejero, P. Vera, Genes encoding acidic and basic class IIIb-1,3-glucanases are expressed
in tomato plants upon viroid infection, Plant Mol. Biol. 24 (1994) 725 – 732.
[20] G. Payne, E. Ward, T. Gaffney, P. Ahl-Goy, M. Moyer, A. Harper, F. Meins, J. Ryals, Evidence for a third structural class of b-1,3-glucanases in tobacco, Plant Mol. Biol. 15 (1990) 797 – 808.
[21] M. Van den Bulcke, G. Bauw, C. Castresana, M. Van Montagu, J. Vandekerckhove, Chraracterization of vac-uolar and extracellularb(1,3)-glucanases of tobacco. Ev-idence for strictly compartmentalized plant defense system, Proc. Natl. Acad. Sci. USA 86 (1989) 2673 – 2677.
[22] Y.J. Kim, B.K. Hwang, Pepper gene encoding a basic pathogenesis-related 1 protein is pathogen and ethylene inducible, Physiol. Plantarum 108 (2000) 51 – 60. [23] H.W. Jung, B.K. Hwang, Isolation, partial sequencing
and expression of pathogenesis-related cDNA genes from pepper leaves infected by Xanthomonas campestris
pv. 6esicatoria, Mol. Plant-Microbe Interact. 13 (2000) 136 – 142.
[24] Y.J. Kim, B.K. Hwang, K.W. Park, Expression of age-related resistance in pepper plants infected with Phy
-tophthora capsici, Plant Dis. 73 (1989) 745 – 747. [25] J.T. Lee, B.K. Hwang, Relation of plant age to bacterial
multiplication in pepper and tomato leaves inoculated with Xanthomonas campestris pv.6esicatoria, Korean J. Plant Pathol. 10 (1994) 18 – 24.
[26] E.S. Kim, B.K. Hwang, Virulence to Korean pepper cultivars of isolates ofPhytophthora capsicifrom differ-ent geographic areas, Plant Dis. 76 (1992) 486 – 489. [27] J.F. Altschul, W. Gish, W. Miller, E.W. Myers, D.J.
Lipman, Basic local alignment search tool, J. Mol. Biol. 215 (1990) 403 – 410.
[28] P. Chomczynski, N. Sacchi, Single-step methods of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction, Anal. Biochem. 162 (1987) 156 – 159.
[29] J. Sambrook, E.F. Fritsch, R. Maniatis, Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY, 1989.
[30] J.K. Hong, H.W. Jung, Y.J. Kim, B.K. Hwang, 2000, Pepper gene encoding a basic class II chitinase is in-ducible by pathogen and ethephon, Plant Science, 159 (2000) 39 – 49.
[31] L.S. Melchers, M.B. Sela-Buurlage, S.A. Vloemans, C.P. Woloshuk, J.S.C. Van Roekel, J. Pen, P.J.M. Van den Elzen, B.J.C. Cornelissen, Extracellular targeting of the vacuolar proteins AP24, chitinase and b-1,3-glucanases in transgenic plants, Plant Mol. Biol. 21 (1993) 583 – 593. [32] M. De Loose, T. Alliotte, G. Gheysen, C. Genetello, J. Gielen, P. Soetaert, M. Van Montagu, D. Inze, Primary structure of a hormonally regulated beta-glucanase of
Nicotiana plumbaginifolia, Gene 70 (1988) 13 – 23. [33] C. Castresana, F. de Carvalho, G. Gheysen, M. Habets,
D. Inze, M. Van Montagu, Tissue-specific and patho-gen-induced regulation of a Nicotiana plumbaginifolia
[35] J.A. van Kan, M.H. Joosten, C.A. Wagemakers, G.C. van den Berg-Velthuis, P.J. de Wit, Differential accumu-lation of mRNAs encoding extracellular and intracellu-lar PR proteins in tomato induced by virulent and avirulent races ofCladosporium ful6um, Plant Mol. Biol. 20 (1992) 513 – 527.
[36] M.M. Chang, L.A. Hadwiger, D. Horovitz, Molecular characteriztion of a pea b-1,3-glucanase induced by
Fusarium solaniand chitosan challenge, Plant Mol. Biol. 20 (1992) 609 – 618.
[37] R. Beffa, F. Meins, Pathogenesis-related functions of plantb-1,3-glucanases investigated by antisense transfor-mation, Gene 179 (1996) 97 – 103.
[38] H. Shinshi, H. Wenzler, J.M. Neuhaus, G. Felix, J. Hofsteenge, F. Meins, Evidence for N- and C-termi-nal processing of a plant defense-related enzyme. Pri-mary structure of tobacco preprob-1,3-glucanase, Proc. Natl. Acad. Sci. USA 85 (1988) 5541 – 5545.
[39] J.F. Bol, H.J.M. Linthorst, B.J.C. Cornelissen, Plant pathogenesis-related proteins induced by virus infection, Annu. Rev. Phytopathol. 28 (1990) 113 – 138.
[40] G.B. Fincher, Molecular and cellular biology associated with endosperm mobilization in germinating cereal grains, Annu. Rev. Plant Physiol. Mol. Biol. 40 (1989) 305 – 346.