Molecular cloning of a soybean class III
b
-1,3-glucanase gene that
is regulated both developmentally and in response to pathogen
infection
Yong Hwa Cheong
a,1, Cha Young Kim
1 a, Hyun Jin Chun
b, Byeong Cheol Moon
c,
Hyeong Cheol Park
a, Jong Kyoung Kim
b, Sung-Ho Lee
b, Chang-deok Han
b,c,
Sang Yeol Lee
a,b, Moo Je Cho
a,b,*
aDepartment of Biochemistry,Gyeongsang National Uni6ersity,Chinju660-701,South Korea
bPlant Molecular Biology and Biotechnology Research Center,Gyeongsang National Uni6ersity,Chinju660-701,South Korea cDepartment of Molecular Biology,Gyeongsang National Uni6ersity,Chinju660-701,South Korea
Received 15 September 1999; received in revised form 14 December 1999; accepted 20 December 1999
Abstract
We isolated and characterized a soybean gene (SGN1) encoding a basic b-1,3-glucanase that is a plant class III isoform of
b-1,3-glucanase. The deduced amino acid sequence of theSGN1 gene is similar to that of the PR-Q%b gene, the basic class III b-1,3-glucanase of tomato. Based on RNA blot hybridization, SGN1 gene expression was detected in all tissues of 4-day old seedlings, but it was present only in root tissue of 30-day old plants. GUS expression analysis carried out in transgenic tobacco plants harboring aSGN1::GUS reporter gene revealed the same expression pattern. Furthermore, the expression of SGN1 was strongly induced by a variety of defense-related signals, such as treatment with H2O2, wounding, or treatment with fungal elicitor
prepared fromPhytophthoraspp as well as inoculation with Pseudomonas syringae. However, the expression level ofSGN1 was hardly induced with jasmonate, ethephon and salicylate. Overall the results suggest that theSGN1 may play a role in both plant development and plant defense against pathogen attack. © 2000 Elsevier Science Ireland Ltd. All rights reserved.
Keywords:Abiotic stresses; Class IIIb-1,3-glucanase; GUS activity; Pathogen infection; Wounding
www.elsevier.com/locate/plantsci
1. Introduction
Plant b-1,3-glucanases (EC 3.2.1.39) are
abun-dant proteins widely distributed among seed plant species. They are involved in diverse physiological
and developmental processes, including
mi-crosporigenesis, pollen germination, fertilization, seed germination and defense against pathogens [1 – 6].
The plant b-1,3-glucanases are divided into at
least three distinct classes depending on their pri-mary structure [1,52]. Class I consists of the basic, vacuolar isoforms which are localized primarily in the epidermis of the lower leaves and in the roots of healthy plants [7]. Class I glucanases undergo substantial post-translational modification includ-ing removal of a carboxyl-terminal extension [8]. Class II comprises three subgroups: (a) acidic, extracellular glucanases (PR-2a, -2b, -2c); (b) closely related isoforms with neutral or basic pI and a carboxyl-terminal extension suggesting a vacuolar localization (GL 153, GL 161), and (c) the stylar-specific extracellular glucanase sp41 [4,9,10]. Class III contains the pathogen-induced glucanase PR-Q, which is an acidic, extracellular
The nucleotide sequence data has been submitted to the EMBL, GenBank™ Nucleotide Sequence Databases under the accession number U41323
* Corresponding author. Tel.: +82-591-7515957; fax: + 82-591-7599363.
E-mail address:[email protected] (M.J. Cho)
1The first two authors made equal contribution to this work.
protein, and it differs by ca. 43% in the sequence from class I and II proteins [9].
The expression of many b-1,3-glucanases can be
induced by fungal elicitors, wounding, salicylic acid (SA), ethylene, and other chemical inducers
[10 – 14].b-glucanase genes are also expressed
dur-ing the hypersensitive response (HR) in
TMV-in-oculated leaves [10,11,15]. b-1,3-glucanases are
thought to act against pathogen directly by digest-ingb-1,3-glucans in fungal cell walls as well as act indirectly by digesting fungal and host polysaccha-rides to produce elicitors capable of evoking the
HR [16 – 19]. Also, the b-1,3-glucanases, especially
in conjunction with chitinase, are capable of hy-drolyze fungal cell walls in vitro [20]. Since both enzymes are co-induced in response to fungal at-tack, their function is believed to be the inhibition of fungal infection [21]. Recently, it has been
reported that class I b-1,3-glucanase deficient
to-bacco plants generated by antisense transforma-tion exhibit decreased susceptibility to necrotic virus infection [22].
So far, class I and II b-1,3-glucanases have been
well studied in terms of gene regulation and
func-tion in plant. However, class III b-1,3-glucanases
have been less studied in their gene regulation
except for PR-Q%a and PR-Q%b, the acidic and
basic class III glucanase from tomato, which are expressed upon virus infection and ethylene
treat-ment, respectively [23]. In particular, cis-acting
elements in the promoters of the class III b
-1,3-glucanases have not yet been characterized. In order to study gene regulation of a class III glu-canase on the promoter level, we isolated a
soy-bean (Glycine max cv Williams) basic class III
b-1,3-glucanase gene, designated as SGN1. In this
paper, we report the isolation and characterization
of SGN1 which is very closely related to the
PR-Q%b gene that encodes a tomato basic class III
b-1,3-glucanase gene. We analyzed the expression
of the SGN1 gene during development and in
response to pathogen signals. Expression ofSGN1
was induced by pathogen signals, such as H2O2,
fungal elicitor prepared from Phytophthora spp
and wounding, but not by ethephon, jasmonic acid (JA) and salicylic acid (SA). These results indicate
that SGN1, a class III basic glucanase, responds
differently to defense signals and is, therefore, differently regulated in comparison class I and II plant glucanases.
2. Materials and methods
2.1. Plant material and treatments
Soybean (G. max cv. Williams) and tobacco
(Nicotiana tabacum cv Xanthi) were used. To ob-tain seedlings, soybean seeds were germinated and grown in darkness for 1 – 5 days in moist vermi-culite at 28°C. To obtain tissue from mature plants, soybean seeds were planted in sterilized soil and grown in a growth chamber (Conviron E15) with a 14 h light (28°C) and 10 h dark (20°C) cycle for around 30 days. All plants were watered to saturation daily and fertilized weekly with 20-20-20 fertilizer (Peters, Allentown, PA)
To analyze expression of the soybean SGN1
gene during development, hypocotyls of soybean seedlings were used. To study stress induction, 4-day old soybean seedlings were exposed to
vari-ous external stresses; sterile-distilled H2O, 0.01%
(V/V) ethephon, 1 mM salicylate (SA), 0.2 mM
jasmonate (JA), 1 mM H2O2, fungal elicitor
pre-pared from Phytophthora spp (50 mg/ml glucose
equivalents) and inoculation of Pseudomonas sy
-ringae pv glycinea (106 cells/ml). The chemicals or
elicitor were sprayed to the soybean seedlings in plastic boxes and incubated for 24 h in the boxes containing 100 ml of the same treatment solutions. For the wounding stress, the 4-day old soybean seedlings were punctured with a 20-gauge needle at 1 mm intervals along opposite sides of the entire seedling. Fungal elicitor had been isolated as de-scribed by Simmons et al. [24] and stored at 4°C until use. For control, total RNA was isolated from water-treated seedlings. After treatment, the
tissue was frozen in liquid N2 and stored at −
80°C until use.
Seeds of transgenic tobacco plants (Ro) were obtained by self pollination of the transgenic plants. The R1 progeny were aseptically germi-nated on wet filter paper discs in petri dishes under a 16 h-light/8 h-dark cycles.
2.2. Cloning of the SGN1 gene
A lambdaEMBL3 soybean genomic library was
screened with the tobacco b-1,3-glucanasegn1 as a
probe [25]. Approximately 300 000 recombinant plaques were screened by plaque hybridization at
60°C in a solution containing 6×SSC (1×SSC is
5×Denhart’s solution [1×Denhart’s solution is
0.02% (w/v) Ficoll 400, 0.02% (w/v)
Polyvinylpyrrolidone, 0.02% (w/v) BSA], 0.5% (w/
v) SDS, 5 mM EDTA and 100 mg/ml sheared,
denatured calf thymus DNA. The membranes
were washed twice in 2×SSC, 0.1% SDS and for
a final washed in 0.2×SSC, 0.1% SDS for 15 min
at 60°C and then exposed to Kodak XAR-5X-ray
film at −80°C. Positive clones were mapped with
restriction enzymes and subcloned into the
pBluscript II vector for DNA sequencing.
2.3. DNA sequencing and analysis
Double-stranded DNA was sequenced by the
dideoxy chain termination method [27] with aTaq
Dye primer cycle sequencing kit using an Applied Biosystem 373A automatic DNA sequencer (ABI). Nucleotide and amino acid sequence analyses were performed with the Macintosh DNASIS program
(Hitachi software Engineering America, San
Bruno, CA).
2.4. Primer extension analysis
To determine the transcriptional start site on the
SGN1 gene, a primer extension experiment was
performed as described by Sambrook et al. [26]
using a synthetic 20-mer oligonucleotide (5%
-gatg-gaagaacttttcccac-3%) complementary to nucleotides
+97 to +77 of the SGN1 gene.
2.5. Genomic Southern blot analysis
Southern blot analysis was carried out as de-scribed by Hong et al. [28] using total soybean DNA isolated by a CTAB precipitation method [29] from etiolated soybean hypocotyls. Hybridiza-tion and washing condiHybridiza-tions were as described above for the library screening.
2.6. RNA isolation and Northern blot analysis
Total RNA was isolated as described by Hong
et al. [28]. Total RNA (20 mg) was denatured and
separated by electrophoresis on a 1.2% agarose-formaldehyde gel, and transferred onto a GeneS-creen Plus membrane (NEN). Hybridization and washing conditions were as described above for the library screening.
2.7. Construction of a SGN1::GUS fusion gene and plant transformation
For the construction of a SGN1::GUS fusion
chimeric gene, a 1.7 kb fragment (with HincII and
HindIII restriction sites on either end) containing
the promoter region and part of the coding region of the SGN1 gene was amplified by PCR using
synthetic oligonucleotide primers [5%
-GTCGAC-TAAGTCCATT-3% (−1618 to −1602) and
5%-GATGGAAGAAGCTTT-TCCCAC-3% (+97 –
+77). The mismatches (bold) were introduced to
create the HincII and HindIII sites (underlined)].
The PCR product was then digested with HincII
and HindIII and subcloned between theSmaI and
HindIII site of pBluescript SK(+). The resulting
recombinant plasmid, referred to pBS-PCR, thus
contained about 1.7 kb of SGN1 promoter. A
BamHI/HincII fragment (−1618 –+97) from
pBS-PCR was then inserted into the BamHI/SmaI
site of the GUS expression vectorpBI101.1 to give
SGN1::GUS construct [30].
The construct SGN1::GUS was introduced into
Agrobacterium LBA4404 by electroporation, and
transgenic tobacco plants were generated by the leaf disc method [31,32]. Transformed plants were selected on Murashige and Skoog (MS) basal
medium [33] containing 200 mg/ml kanamycin and
500mg/ml carbenicillin and grown at 25°C under a
16 h-light/8 h-dark cycle.
2.8. Detection of GUS acti6ity
To test for expression of the SGN1::GUS
con-struct during seed germination, transformed to-bacco seeds were sterilized and allowed to germinate for 1 or 4 days. To test for induction of
SGN::GUS gene expression, leaves from the 6- or
7-week old transformed plants were used after wounding, treatment with various stress chemicals,
such as SA, JA, H2O2, and ethephon, for 18 h or
inoculation with Pseudomonas syringae pv tabaci
(Pst) for 36 h. The tests were done by fluorometric and histochemical analysis.
Fluorometric GUS assays were carried out as described by Jefferson et al. [30]. Protein concen-tration was determined with a Bradford assay [34], and GUS activity was expressed as nmol 4-MU produced per mg protein per min.
microscopically viewed and photographed with a Leitz stereomicroscope.
3. Results
3.1. Isolation and characterization of the soybean basic class III b-1,3-glucanase (SGN1) gene
While studying the overall organization and
reg-ulatory mechanisms of soybean b-1,3-glucanase
genes, a soybean genomic library was screened by
plaque hybridization using a tobacco b
-1,3-glu-canase (gn1) gene as a probe [25]. One positive
phage containing a 15 kb insert was isolated and characterized by hybridization, restriction map-ping, and sequence analysis. A putative soybean
b-1,3-glucanase (SGN1) gene, on a 5.3 kb AccI/
HindIII DNA fragment was isolated and
sub-cloned. The complete nucleotide sequence of the
SGN1 gene with its 5%- and 3%-flanking regions is
determined (accession number U41323). The
cod-ing region of theSGN1 gene consists of two exons
(94 and 953 bp) separated by one intron (1981 bp). It is thus expected to yield a precursor protein of 348 amino acids. The transcription start site for
the SGN1 gene was determined using primer
ex-tension analysis (data not shown). RNA prepared from 4-day old soybean seedlings was hybridized to an oligonucleotide complementary to the
cod-ing strand of the SGN1 gene. After extension
using reverse transcriptase, two products differing in length by two nucleotides were observed. The
calculated ends of the two 5% untranslated leaders
on these two mRNA species were 58 (C base) and 56 (T base) nucleotides upstream of the translation start site codon (ATG). The start site of the longer
leader was designated as +1. A putative TATA
box and a CAAT-like sequence are found at
posi-tions −35 to −30 and −66 to −62,
respec-tively. At the 3%-end of the gene, a consensus
sequences (AATAAA) for a polyadenylation sig-nal is found at position 3236 to 3241 bp.
In order to determine the copy number of the
b-1,3-glucanase genes in the soybean genome,
ge-nomic Southern blot analysis was carried out
us-ing the EcoRI/HindIII fragment (coding region)
of the genomic clone,SGN1, as a probe. Only one
or two hybridizing bands were detected over a range of restriction endonuclease digestions (data not shown). These results therefore indicate that
the SGN1 gene is likely present in the soybean
genome as a single or at most two-copy gene.
3.2. SGN1 resembles other plant class III glucanases
The deduced amino acid sequence of the SGN1
gene contains 348 amino acids, with a putative
signal peptide of 33 amino acids at the N
-termi-nus. The calculated molecular mass is 38.2 kDa, whereas that of the mature peptide is 34.6 kDa. The isoelectric point (pI) of the total peptide was calculated as 9.08. The predicted amino-terminal sequence of the mature peptide, G-S- (nucleotide position 2138 to 2143), is the same as that found
in tobacco or tomato b-1,3-glucanases [23,25].
Two consensus sequences for N-glycosylation
(N-X-S/T) are located at amino acid positions
199-201 (N-N-T) and 220 – 222 (N-I-S). There are no C-terminal extension sequences, which are found in dicot vacuolar class I basic b-1,3-glucanases [8]. The amino acid sequence of SGN1 is 70 and 65% identical to tobacco PR-Q [9] and tomato PR-Q%b [23], respectively, which are basic class III
b-1,3-glucanases (Fig. 1). Tomato PR-Q%a, an
acidic class III b-1,3-glucanase [23], shares 55%
identity with SGN1 and tobacco GN1 [25], a basic
class I b-1,3-glucanase and tobacco GN2 [10], an
acidic class II tobacco b-1,3-glucanase, are 50 and
45% identical, respectively. Plant b-1,3-glucanase
sequences have been divided into three major groups, including basic class I, acidic class II and class III isoforms [1]. Based on our data, SGN1 has to be classified as a plant class III isoform.
3.3. De6elopmental expression of the SGN1 gene in soybean seedlings
In order to investigate the developmental
stage-specific expression of the SGN1 gene in soybean
seedlings, we performed northern blot analysis of total RNA isolated from hypocotyls of 1-, 2-, 3-, 4-, and 5-day old etiolated seedlings using the
coding region of the SGN1 gene (EcoRI-HindIII
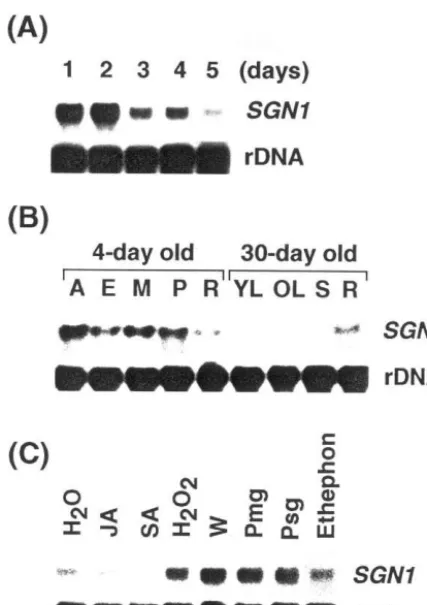
fragment) as probe. Fig. 2A shows the change in
SGN1 mRNA levels in hypocotyls. The mRNA
To examine spatial distribution pattern of the
SGN1 mRNA, we dissected dark-grown 4-day old
seedlings and light-grown 1-month old green plants, as described previously [28]. As shown in
Fig. 2B, theSGN1 transcripts were abundant in all
tissues of dark-grown 4-day old seedlings, such as the apical, elongating and mature sections of the hypocotyl, plumule, and root. However, in the light-grown 1-month old green plants, the mRNA was only abundant in the root tissue. The tran-script was hardly detected in the leaves and stems of 1-month old green plants. Thus, expression of
the SGN1 gene is developmentally regulated in
soybean.
3.4. Accumulation of SGN1 mRNA in response to
wounding and defense-related signals
b-1,3-glucanase genes in several plant species
have been shown to be induced in response to various treatments [1]. To determine whether the
expression of the SGN1 gene can be stimulated by
external stimuli, 4-day old soybean seedlings were treated with various stress agents for 24 h and the transcript level of the gene was assayed by RNA
gel blot analysis. As shown in Fig. 2C,
transcrip-tion ofSGN1 was induced in response to a variety
of stress treatments such as wounding, exposure to
H2O2, fungal elicitor prepared from Phytophthora
parasitica (Pmg), and Pseudomonas syringae pv
glycinae (Psg), an avirulent pathogen of soybean. However, treatments with jasmonate (JA), ethep-hon and salicylate (SA), compounds which report-edly induce the expression of other PR proteins
[35,36], did not induce the expression of theSGN1
gene.
3.5. Construction and analysis of the expression of
SGN1::GUS fusion protein under control of
SGN1 promoter
To facilitate the study of the regulatory
charac-teristics of the SGN1 gene promoter, an
SGN1::GUSchimeric gene was constructed by
fus-ing 1.6 kb of the promoter region of the SGN1
gene to the coding region of the bacterial b
-glu-curonidase (GUS) gene (Fig. 3A). The construct was introduced into tobacco cells, and kanamycin-resistant transgenic tobacco plants were generated, and analyzed for GUS activity.
Fig. 1. Comparison of the soybean class III glucanase (SGN1) amino acid sequence with other plant b-1,3-glucanases. Shaded areas indicate identical amino acids with the SGN1 sequence, and gaps were introduced for optimal alignment. The end of the signal peptide is marked with an arrow pointing out an expected processing site between alanine and glutamine. Tob PR-Q, tobacco basic class IIIb-1,3-glucanase [9]; Tom PR-Q%b, tomato basic class IIIb-1,3-glucanase [23]; Tom PR-Q%a, tomato acidic
Fig. 2. Northern blot analysis. RNA blot showing stage-spe-cific expression of the SGN1 gene in 1 to 5-day old soybean seedlings after germination. The membrane was exposed to the X-ray film at−80°C for 1 day. (B) Twenty microgram of total RNA isolated from apical (A), elongating (E), mature (M) sections of the hypocotyl, plumule (P) and roots (R) of dark-grown 4-day old soybean seedlings. RNA samples of young leaves (YL), old leaves (OL), stems (S), and roots (R) were obtained from light-grown 30-day old soybean plants. The membrane was exposed to the X-ray film at −80°C for 2 days. (C) Effect of various defense signaling molecules on the expression of the SGN1 gene. Total RNA was isolated from 4-day old soybean seedlings treated with JA (0.2 mM), SA (1 mM), H2O2(1 mM), W (wounding), ethephon (0.01%),
Pmg (50mg/ml of fungal elicitor prepared fromPhytophthora parasitica var. nicotianae) and Psg (106 cells/ml of Pse
-domonas syringae pv glycinea) for 24 h. Solutions of ethep-hon, salicylic acid, H2O2and elicitor were prepared in water
and jasmonate was prepared in ethanol. For control, total RNA was isolated from water-treated seedlings. The mem-brane was exposed to the X-ray film at −80°C for 1 day.
lation with Pseudomonas syringae pv tabaci (Pst),
but not by salicylate, jasmonate and ethephon. These were the same results as obtained by north-ern blot analysis.
Fig. 3. Analysis of stress-induced expression of GUS activity in transgenic tobacco plants containing theSGN1::GUS con-struct. (A)SGN1::GUSfusion construct. A 1618 bp fragment of flanking region extending 5% from the transcription start
site of SGN1 was fused to the coding region of a GUS reporter gene in pBI101.1. The arrow indicates the direction of the transcription of theSGN1 andGUSgene as driven by theSGN1 promoter. (B) Analysis of stress-induced expression of GUS activity in transgenic tobacco plants containing the
SGN1::GUS construct. GUS activity was determined for wa-ter-treated (control) leaf tissue of transgenic plants and for leaf tissue that had been treated by floating them on solutions of the various stress signal substances, such as ethephon, salicylate, fungal elicitor ofPhytophthora parasiticavar. nico-tianae (Pmg), H2O2and jasmonic acid for 24 h. Some
treat-ments consisted of wounding or inoculation with
Pseudomonas syringae pvtabaci (Pst). Average GUS activity was derived from five independent transformants. The num-bers on the right indicate x-fold levels of GUS activity induced by the various treatments relative to the GUS activity of the water treated control (=1). (C) Analysis of the GUS activity in tissues of different developmental stages. The data represent average GUS activities obtained from six indepen-dent transgenic plants examined approximately 7 weeks after germination. GUS activity was determined in different plant regions designated I to V as demonstrated by the diagram at the left.
To examine whether external stresses have any
effect on the expression of theGUSgene under the
control of theSGN1 promoter, we looked at GUS
activity in fully expanded leaves of transgenic
plants containing the SGN1::GUS reporter gene
after stress treatments. As shown in Fig. 3B,
ex-pression of the SGN1::GUS gene was induced by
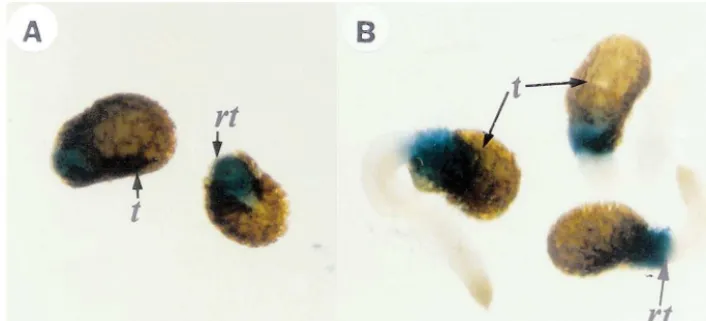
inocu-Fig. 4. Histochemical localization of GUS activity in germinatingSGN1::GUS transgenic tobacco seeds. The transgenic plants containing the introducedSGN1::GUSgene were grown from seeds on MS agar plates for 1- (A) or 4- days (B). The plants were stained with X-Gluc. After staining the plants were treated with ethanol to remove chlorophyll: t, testa; rt, root.
Fig. 5. Histochemical localization of GUS activity in transgenic tobacco leaves after bacterial inoculation and wounding. Transgenic plants containing theSGN1::GUS construct were treated after approximately 4 weeks of growth on MS agar plates. Tobacco leaves were locally inoculated with mock (control) (A) and with aPsedomonas syringaepvtabacistrain carrying avr C (B) for 24 h. A leaf treated by wounding is shown in (C). After staining with X-Gluc, the leaves were placed in ethanol to stop the reaction and to remove chlorophyll.
To determine whether theSGN1::GUSconstruct
is developmentally regulated, we analyzed six inde-pendent transgenic plants which had grown in the greenhouse for approximately 8 weeks after
trans-fer to soil. The SGN1::GUS transgenic plants
showed higher GUS activity in the roots and lower leaves than upper leaves (Fig. 3C). There was actually a gradient of GUS activity seen through-out the plant with maximum values in the roots, whereas very low activity was present in the upper leaves of the plant. Our data, therefore, suggest
that SGN1 expression is differentially regulated at
developmental stages and in response to pathogen attack.
3.6. Spatial localization of SGN1::GUS
expression
To better understand the developmental
regula-tion of the SGN1 gene activity and its tissue
specificity, expression patterns of the chimeric
SGN1 gene were analyzed in healthy transgenic
plants using histochemical GUS staining (Fig. 4). During early seedling growth (3 – 4 days after
sow-ing), SGN1::GUS gene expression was detected in
To further identify the plant cells that expressed
GUS under the control of SGN1::GUS construct
during a defense response, the GUS activity was histochemically assayed 24 h after local infiltration ofP. syringaepvtabaci(106cells/ml) and
mechan-ical wounding to the transgenic plants, respec-tively. GUS activities were immediately detected at high levels in the tissue surrounding the local lesions, which were developed by both bacterial infiltration and mechanical wounding (Fig. 5).
4. Discussion
In this paper, we describe SGN1, a gene from
soybean that encodes a basic class III b
-1,3-glu-canase. The structure of the gene, the tissue-spe-cific and developmental-spetissue-spe-cific expression of the gene in the healthy plants, and the expression pattern of the gene in response to various stresses and to signaling molecules were characterized. In addition, a chimeric gene was constructed by
fus-ing the SGN1 promoter to a GUS reporter gene,
and the expression of the chimeric gene was stud-ied in transgenic tobacco plants.
Since the deduced amino acid sequence of the SGN1 shares 70% identity with the tobacco basic
class III b-1,3-glucanase, PR-Q, [9] and 65%
iden-tity with the tomato basic b-1,3-glucanase,
PR-Q%b, [23], SGN1 can be classified as a plant basic
class III b-1,3-glucanase (Fig. 1).
In young soybean seedlings 4 days after
germi-nation, the transcript levels of SGN1 gene were
high in all tissues. However, the accumulation of
SGN1 transcript was quite low in the leaves and
stems of 30-day old plants, while high in the root
tissues indicating spatial/temporal regulation of
SGN1 expression (Fig. 2). Recently, Jin et al.
(1999) reported similar results that the mRNA
levels of all 12 classes of b-1,3-glucanase genes
from soybean were low in young leaves, whereas
SGlu2, SGlu4, SGlu7, and SGlu12 mRNA were
highly accumulated in young roots and hypocotyls [52]. Transgenic tobacco plants harboring the
GUS reporter gene driven by the SGN1 promoter
also showed high levels of GUS activity in the roots of both young seedling and old plants, while low levels of GUS activity were detected in stems
and leaves. Thus the SGN1 promoter was highly
active in the lower part of transgenic tobacco plants decreasing in the upper plant parts which
correlates with the age of the leaves and the stem sections (Fig. 3). This overall expression pattern of
SGN1 is similar to plant class I b-1,3-glucanases
[14,25,37].
The class I glucanases are expressed in many tissues and are regulated by both phytohormones and salicylic acid [38 – 40] as well as in response to stresses such as wounding, microbial infection and UV irradiation [41,42]. Recently, it was reported that a tobacco class I glucanase was down regu-lated by ABA treatment [43]. The acidic, extracel-lular class II glucanases are expressed in the floral organs of healthy plants and in newly germinated seedlings. Class II glucanases also appear locally around necrotic viral lesions and systemically in leaves after inoculation with TMV (tobacco mo-saic virus) and upon treatment with salicylate [37].
It has been reported that b-1,3-glucanases are
involved in controlling the size of lesions and the spread of pathogen [22].
A variety of defense-related signals including wounding, H2O2, the fungal elicitor fromPhytoph
-thora parasitica (Pmg), and the avirulent bacterial
pathogen, Pseudomonas syringae pv glycinea (Psg)
lead to the accumulation of SGN1 transcript.
However the expression of the gene was barely induced upon ethephon, salicylate and jasmonate treatments, although these substances have been
shown to regulate other b-1,3-glucanases (Fig. 2).
We obtained similar responses from the transgenic
tobacco plants containing the SGN1::GUS
re-porter gene construct (Fig. 4). Furthermore, the
expression of SGN1 was highly induced around
the necrotic lesions when evoking a hypersensitive
response of the plants by infection ofPseudomonas
syringaepv tabaci(Pst) (Fig. 5). This response was similar to the induction of many other plant de-fense-related genes. These results indicate that
soy-bean class III b-1,3-glucanase, SGN1, may be
involved in plant defense and developmental mechanisms.
Severalcis-acting elements that are required for
ethylene- and SA- inducible expressions of b-1,3
glucanase genes have been identified in plants
[35,44]. However, the SGN1 promoter does not
contain the SA or ethylene-inducing cis element.
This was supported by our finding that the SGN1
gene expression was not induced by SA or
ethep-hon treatment (Figs. 2 and 3). The SGN1
pro-moter, though, contains putative cis-acting
regula-tion in PR proteins including putative fungal elici-tor responsive elements such as the TAATTG box and the EIRE (GTCAG) box. Four putative
TAATTG elements are located at positions −
1006 to −993 (reverse), −816 to −803 (reverse),
−776 to −763 and −194 to −181 bp in the
SGN1 promoter. The TAATTG element has been
reported to be present in the promoter region of
many plant genes. It reportedly is a cis element
required for fungal elicitor-mediated expression of
the parsley pr2 gene [45]. Other fungal elicitor
responsive elements, such as the GTCAG motif, have been found for several defense-related genes
[46 – 50]. TheSGN1 promoter contains three
puta-tive EIRE elements: between positions −638 to
−622, −281 to 289 bp and −105 to −89 bp.
The two fungal elicitor elements, the TAATTG box and the EIRE box, are probably required for
the defense-related regulation of theSGN1 gene. A
putative bHLH box (CAXXTG), linked to wound inducible expression [51], is also located in the
SGN1 promoter. Further work will be needed to
characterize the all cis-elements related to defense
responses.
Acknowledgements
This work was supported by the Science and Technology Policy Institute grant (BT-3-2-04) and G7 grant (99-g-08-02-06) to M.J. Cho, and grant from the KOSEF to the Plant Molecular Biology and Biotechnology Research Center (PMBBRC).
References
[1] F. Meins Jr, J.-M. Neuhaus, C. Sperisen, J. Ryals Jr, The primary structure of plant pathogenesis-related glu-canohydrolases and their genes, in: T. Boller, F. Meins Jr (Eds.), Genes Involved in Plant Defense, Springer-Ver-lag, Berlin, 1992, pp. 245 – 282.
[2] P.A. Bucciaglia, A.G. Smith, Cloning and characteriza-tion ofTag1, a tobacco antherb-1,3-glucanase expressed during tetrad dissolution, Plant Mol. Biol. 24 (1994) 903 – 914.
[3] H.D. Roggen, R.G. Stanley, Cell-wall hydrolysing en-zymes in wall formation as measured by pollen-tube extension, Planta 84 (1969) 295 – 303.
[4] N. Ori, G. Sessa, T. Lotan, S. Himmelhoch, R. Fluhr, A major stylar matrix polypeptide (sp41) is a member of the pathogenesis-related proteins superclass, EMBO J. 9 (1990) 3429 – 3436.
[5] G. Leubner-Metzger, C. Frundt, R. Vogeli-Lange, F. Meins Jr, Class I b-1,3-glucanase in the endosperm of tobacco during germination, Plant Physiol. 109 (1995) 751 – 759.
[6] R. Vogeli-Lange, C. Frundt, C.M. Hart, R. Beffa, F. Nagy, F. Meins Jr, Evidence for a role ofb-1,3-glucanase in dicot seed germination, Plant J. 5 (1994) 273 – 278. [7] D. Keefe, U. Hiinz, F. Meins Jr, The effect of ethylene
on the cell type-specific and intracellular localization of
b-1,3-glucanase and chitinase in tobacco leaves, Planta 182 (1990) 43 – 51.
[8] H. Shinshi, H. Wenzler, J.-M. Neuhaus, G. Felix, J. Hofstoenge, F. Meins Jr, Evidence for N- and C-termi-nal processing of a plant defense-related enzyme: primary structure of tobacco prepro-b-1,3-glucanase, Proc. Natl. Acid. Sci. USA 85 (1988) 5541 – 5545.
[9] G. Payne, E. Ward, T. Gaffney, P.A. Goy, M. Moyer, A. Harper, F. Meins Jr, J. Ryals, Evidence for a third structural class ofb-1,3-glucanase in tobacco, Plant Mol. Biol. 15 (1990) 797 – 808.
[10] E. Ward, G. Payne, M. Moyer, S. Williams, S. Dincher, K. Sharkey, J. Beck, H. Taylor, P.A. Goy, F. Meins Jr, J. Ryals, Differential regulation ofb-1,3-glucanase mes-senger RNAs in response to pathogen infection, Plant Physiol. 96 (1991) 390 – 397.
[11] F. Cote, J.R. Cutt, A. Asselin, D.F. Klessig, Pathogene-sis-related acidic b-1,3-glucanase genes of tobacco are regulated by both stress and developmental signals, Mol. Plant-Microbe Interact. 4 (1991) 173 – 181.
[12] G. Delp, E.T. Palva, A novel flower-specific Arabidopsis gene related to both pathogen-induced and developmen-tally regulated plant b-1,3-glucanase genes, Plant Mol. Biol. 39 (1999) 565 – 575.
[13] J. Malamy, J. Hennig, D.F. Klessig, Temperature-depen-dent induction of salicylic acid and its conjugates during the resistance response to tobacco mosaic virus infection, Plant Cell 4 (1992) 359 – 366.
[14] A.D. Neale, J.A. Wahleithner, M. Lund, H.T. Bonnett, A. Kelly, D.R. Meeks-Wagner, W.J. Peacock, E.S. Den-nis, Chitinase, b-1,3-glucanase, osmotin and extension are expressed in tobacco explants during flower forma-tion, Plant Cell 2 (1990) 673 – 684.
[15] H.J. Linthorst, L.S. Melchers, A. Mayer, J.S. van Roekel, B.J.C. Cornelissen, J.F. Bol, Analysis of gene families encoding acidic and basic b-1,3-glucanase of tobacco, Proc. Natl. Acid. Sci. USA 87 (1990) 8756 – 8760.
[16] F. Mauch, L.A. Staehelin, Functional implication of the subcellular localization of ethylene-induced chitinase and
b-1,3-glucanase in bean leaves, Plant Cell 1 (1989) 447 – 457.
[17] M.M. Chang, L.A. Hadwiger, D. Horovitz, Molecular characterization of a pea b-1,3-glucanase induced by
Fusarium solaniand chitosan challenge, Plant Mol. Biol. 20 (1992) 609 – 618.
[18] Q. Zhu, E.A. Maher, S. Masoud, R.A. Dixon, C. Lamb, Enhanced protection against fungal attack by constitu-tive coexpression of chitinase and glucanase genes in transgenic tobacco, Bio/Technology 12 (1994) 807 – 812. [19] J. Ebel, D. Scheel Jr, Elicitor recognition and signal
Involved in Plant Defense, Springer-Verlag, Vienna/New York, 1992, pp. 183 – 205.
[20] F. Mauch, B. Mauch-Mari, T. Boller, Antifungal hydro-lases in pea tissue. II. Inhibition of fungal growth combi-nation of chitinase and b-1,3-glucanases, Plant Physiol. 88 (1988) 936 – 942.
[21] U. Vogeli, F. Meins Jr, T. Boller, Coordinated regulation of chitinase and b-1,3-glucanase in bean leaves, Planta 174 (1988) 364 – 372.
[22] R.S. Beffa, R.-M. Hofer, M. Thomas, F. Meins Jr, Decreased susceptibility to viral disease of b -1,3-glu-canase-deficient plants generated by antisense transfor-mation, Plant Cell 8 (1996) 1001 – 1011.
[23] C. Domingo, V. Conejero, P. Vera, Genes encoding acidic and basic class III b-1,3-glucanases are expressed in tomato plants upon viroid infection, Plant Mol. Biol. 24 (1994) 725 – 732.
[24] C.R. Simmons, J.C. Litts, N. Huang, R.L. Rodriguez, Structure of a rice b-glucanase gene by ethylene, cy-tokinin, wounding, salicylic acid and fungal elicitors, Plant Mol. Biol. 18 (1992) 33 – 45.
[25] C. Castresana, F. de Carvalho, G. Gheysen, M. Habets, D. Inze, M. Van Montagu, Tissue-specific and pathogen-induced regulation of a Nicotiana plumbaginifolia b -1,3-glucanase gene, Plant Cell 2 (1990) 1131 – 1143.
[26] J. Sambrook, E.F. Fritsch, T. Maniatis, Molecular cloning: A Laboratory Manual, second ed, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, 1989.
[27] F. Sanger, S. Nicklen, A.R. Coulson, DNA sequencing with chain-terminating inhibitors, Proc. Natl. Acid. Sci. USA 74 (1977) 5463 – 5467.
[28] J.C. Hong, R.T. Nagao, J.L. Key, Characterization and sequence analysis of a developmentally regulated puta-tive cell wall protein gene isolated from soybean, J. Biol. Chem. 262 (1987) 8367 – 8376.
[29] M.G. Murray, W.F. Thompson, Rapid isolation of high molecular weight plant DNA, Nucl. Acid Res. 8 (1980) 4321 – 4325.
[30] R.A. Jefferson, T.A. Kavanagh, M.W. Bevan, GUS fu-sions: b-glucuronidase as a sensitive and versatile gene fusion marker in higher plants, EMBO J. 6 (1987) 3901 – 3907.
[31] W.J. Dower, J.F. Miller, C.W. Ragsdale, High efficiency transformation of Escherichia coli by high voltage elec-troporation, Nucl. Acids. Res. 16 (1988) 6127 – 6145. [32] R.B. Horsch, J.E. Fry, N.L. Hoffmann, D. Eichholfz,
S.G. Rogers, R.T. Fraley, A simple and general method for transferring genes into plants, Science 227 (1985) 1229 – 1231.
[33] T. Murashige, F. Skoog, A revised medium for rapid growth and bioassays with tobacco tissue cultures, Phys-iol. Plant. 15 (1962) 473 – 497.
[34] M.M. Bradford, A rapid and sensitive method for the quantitation of protein utilizing the principle of protein-dye binding, Anal. Biochem. 72 (1976) 248 – 254. [35] J. Shah, D.F. Klessig, Identification of a salicylic
acid-re-sponsive element in the promoter of the tobacco patho-genesis-related b-1,3-glucanase gene, PR-2d, Plant J. 10 (1996) 1089 – 1101.
[36] M.D. Van de Rhee, J.A. Van Kan, M.T. Gonzalez-Jaen, J.F. Bol, Analysis of regulatory elements involved in the induction of two tobacco genes by salicylate treatment and virus infection, Plant Cell 2 (1990) 357 – 366. [37] J. Hennig, R.E. Dewey, J.R. Cutt, D.F. Klessig,
Patho-gen, salicylic acid and developmental dependent expres-sion of b-1,3-glucanase/GUS gene fusion in transgenic tobacco plants, Plant J. 4 (1993) 481 – 493.
[38] M. De Loose, T. Alliotte, G. Gheysen, C. Genetello, J. Gielen, P. Soetaert, M. Van Montagu, D. Inze, Primary structure of a hormonally regulatedb-glucanase ofNico
-tiana plumbaginifolia, Gene 70 (1988) 13 – 23.
[39] G. Felix, F. Meins Jr, Ethylene regulation of b -1,3-glu-canase in tobacco, Planta 172 (1987) (1987) 386 – 392. [40] J. Memelink, H.J. Linthorst, R.A. Schilperoort, J.H.
Hoge, Tobacco genes encoding acidic and basic isoforms of pathogenesis-related proteins display different expres-sion patterns, Plant Mol. Biol. 14 (1990) 119 – 126. [41] F.T. Brederode, H.J. Linthorst, J.F. Bol, Differential
induction of acquired resistance and PR gene expression in tobacco by virus infection, ethephone treatment, UV light and wounding, Plant Mol. Biol. 17 (1991) 1117 – 1125.
[42] M. Ohme-Takagi, H. Shinshi, Structure and expression of a tobacco b-1,3-glucanase gene, Plant Mol. Biol. 15 (1990) 941 – 946.
[43] E. Rezzonico, N. Flury, F. Meins, R. Beffa, Transcrip-tional down-regulation by abscisic acid of pathogenesis-related b-1,3-glucanase genes in tobacco cell cultures, Plant Physiol. 117 (1998) 585 – 592.
[44] E. Alonso, F. de Carvalho Niebel, P. Obregon, G. Gheysen, D. Inze, M. Van Montagu, C. Castresana, Differential in vitro DNA binding activity to a promoter element of the gn1 b-1,3-glucanase gene in hypersensi-tively reacting tobacco plants, Plant J. 7 (1995) 309 – 320. [45] U. Korfhage, G.F. Trezzini, I. Meier, K. Hahlbrock, I.E. Somssich, Plant homeodomain protein involved in tran-scriptional regulation of a pathogen defense-related genes, Plant Cell 6 (1994) 695 – 708.
[46] D. Raventos, A.B. Jensen, M.B. Rask, J.M. Casacu-berta, J. Mundy, B. San Segundo, A 20 bp cis-acting element is both necessary and sufficient to mediate elici-tor response of a maize PRms gene, Plant J. 7 (1995) 147 – 155.
[47] S.A. Warner, A. Gill, J. Draper, The developmental expression of the asparagus intracellular PR protein (AoPR1) gene correlates with sites of phenylpropanoid biosynthesis, Plant J. 6 (1994) 31 – 34.
[48] C. Despres, R. Subramaniam, D.P. Matton, N. Brisson, The activation of the potato PR-10a gene requires the phosphorylation of the nuclear factor PBF-1, Plant Cell 7 (1995) 589 – 598.
[49] P.J. Rushton, J.T. Torres, M. Parniske, P. Wernert, K. Hahlbrock, I.E. Somssich, Interaction of elicitor-induced DNA binding proteins with elicitor response elements in the promoters of parsley PR genes, EMBO J. 15 (1996) 5690 – 5700.
[50] Y. Fukuda, H. Shinshi, Characterization of a novel
[51] A. Kawaoka, T. Kawamoto, M. Sekine, M. Yoshida, M. Takano, A. Shinmyo, A cis-acting element and atrans -act-ing factor involved in the wound-induced expression of a horseradish peroxidase gene, Plant J. 6 (1994) 87 – 97.
[52] W. Jin, H.T. Horner, R.G. Palmer, R.C. Shoemaker, Analysis and mapping of gene families encoding
b-1,3-glucanases of soybean, Genetics 153 (1999) 445 – 452.