Indonesian Journal of Biotechnology, June, 2016 Vol. 21, No. 1, pp. 1–11
Evaluation of Antimicrobial Activity and Identification of
Yellow Pigmented Marine SpongeAssociated Fungi from Teluk Awur,
Jepara, Central Java
Mada Triandala Sibero
1,2,*, Desy Wulan Triningsih
3, Ocky Karna Radjasa
2,4, Agus
Sabdono
2,4, Agus Trianto
3,4Introduction
Pigments are chemical compounds which give colours because of its ability to absorb light in the wavelength range at visible region (DelgadoVargas et al., 2000). The utilization of pigments is increasing every year due to the increase of demands in food, beverages, pharmaceutical and cosmeceutical industries (Rymbai et al., 2011; Venil et al., 2013). Natural pigments are known to have biological activities as its advantages over the synthetic pigments (Rostami et al., 2016; Normanet al., 2016; Yolmehet al., 2016).
Antimicrobial activity is one of biological activities showed by natural pigments. Salem
et al. (2014) successfully extracted natural pigment from Carthamus tinctorius. The pigments displayed antimicrobial activity against MethicillinResistant Staphylococcus aureus(MRSA),Escherichia coliATCC 25218 andCandida albicans. Another research reported that carotenoids pigment were able to inhibit
Salmonella enteritidis(PTCC 1709) andE. coli
(PTCC1260) (Rostamiet al., 2016). In addition, crude extract of pigment fromStreptomyces
sp. D25 was reported to have antibacterial activity against biofilm forming bacteria such asPseudomonassp. P1.,Bacillussp. P13 and
Alcaligenssp. M28 (Radhakrishnanet al., 2016). Marine microorganisms are known as potential producer of natural pigment with great antibacterial activity. There are several microorganisms are known life associated 1Department of Coastal Resources Management, Faculty of Fisheries and Marine Science,
Diponegoro University, Semarang, Indonesia
2Laboratory of Tropical Marine Biotechnology, Integrated Laboratory of Diponegoro
University, Semarang, Central Java, Indonesia
3Laboratory of Marine Natural Product, Integrated Laboratory of Diponegoro University,
Semarang, Indonesia
4Department of Marine Science, Faculty of Fisheries and Marine Science, Diponegoro
University, Semarang, Indonesia
Abstract
Marine sponge associated fungi are known as potential source of metabolites with various biological activities. Natural pigment is one of metabolite which produced by microorganisms. Several researches reported the antimicrobial activity from natural pigment. Unfortunately there are lack of information about marine fungi natural pigment and its producer. The aims of this research were to identify yellow pigmented Indonesian marine spongeassociated fungi, to extract the pigment, and to study the antimicrobial activity of the pigment against clinical MDR bacteria and clinical pathogenic fungi. Sponge associatedfungus isolate MT23 was successfully identified as Trichoderma parareesei. The fungal pigment could be extracted only in methanol with yield 6.22±0.29%. The pigment could inhibited Salmonella typhi and Escherichia coli MDR strains. The biggest antibacterial activity was shown by concentration 1000µg/mL againstS. typhi with inhibition zone was 4.03±0.06 mm.
Keywords: Associated fungi, pigment,Trichoderma parareesei
*Corresponding author: Mada Triandala Sibero
Faculty of Fisheries and Marine Science, Diponegoro University, Jalan Imam Bardjo, SH., Semarang 50241, Indonesia
with marine organisms especially sponges such as bacteria, actinomycetes and fungi (Idraningrat et al., 2016; Wu et al., 2016). Ibrahim et al. (2014) studied antimicrobial activity of prodigiosin (red pigment) from spongeassociated bacteriaSerratia marcescens
IBRL USM 84. This pigment greatly inhibited MRSA and had good activity inhibitedBacillus cereus, Bacillus licheniformisandAgrobacterium tumefaciens. In 2015, Sureshet al.(2015) observed antibacterial activity of red pigment produced byHalolactibacillus alkaliphilusMSRD1 which isolated from seaweed againstS. aureusand
Salmonella typhi. In addition, Srilekha et al.
(2016) successfully isolated marine pigmented bacteria and extracted the pigment then studied its antimicrobial activity. Result showed the pigment had great antimicrobial activity against bacteria such as S. aureus,
Proteus vulgaris, K. pneumoniae, E. coli, and fungi such asFusariumsp.,C. albicans,Mucor
sp., andA. flavuswith inhibition zone range 18–28 mm. These reports show great potential of natural pigments from marine bacteria as antibacterial agent.
However, there are still a few researches about the potential of natural pigments from marine fungi and its antibacterial activity against clinical multidrug resistant bacteria (MDR) and clinical pathogenic fungi. MDR bacteria are bacteria which already resistant to several antibiotics that should be effective to inhibit the growth of the bacteria (Magiorakos
et al., 2012). This is a big issue for public health. The aims of this research were to identify Indonesian yellow pigmented spongeassociated fungi, to extract the pigment, and to study the antimicrobial activity of the pigment against clinical MDR bacteria and clinical pathogenic fungi.
Materials and Methods
Media and chemicals
This research used Malt Extract Agar (MEA) M137 base with mycological peptone and MacConkey M081B agar from HiMedia Laboratories Pvt. Ltd. (Swastik Disha Business Park, Via Vadhani, Ind. Est., LBS Marg, Mumbai400086, India), Nutrient Agar CM0003
from Oxoid Ltd. (Basingstoke, Hampsphire, England), Mueller Hinton Agar (MHA) ACC, to CLSI 1.05435.0500 from Merck KGaA (Darmstadt, Germany) while agarose was bought from Promega. Solvents such as methanol, ethyl acetate and nhexane were bought from Merck. DNA extraction used Chelex®100 (SigmaAldrich), PCR master mix GoTaq Green Master Mix from Promega Corporation (2800 Woods Hollow Road·Madison, WI 537115399 U.S.A) while for primer (ITS1 and ITS4) from Macrogen (1330 Piccard Drive Rockville, MD, 20850 United States).
Fungal isolate
Fungus MT23 was culture collection of Marine Fungi Division of Tropical Marine Biotechnology Laboratory, Integrated Laboratory of Diponegoro University, Semarang, Central Java, Indonesia. This fungus was isolated from unidentified marine sponge from Teluk Awur Bay, Jepara, Central Java.
MDR bacteria and clinical pathogenic fungi isolates
MDR bacteria and clinical pathogenic fungi were obtained from Dr. Kariadi General Hospital Medical Center, Semarang, Central Java, Indonesia and Diponegoro National Hospital, Semarang, Central Java, Indonesia. Bacteria E. coli,S. typhiand fungiMalassezia furfurandTrichophyton rubrumwere used in this study.
Fungal culture
MDR bacteria and clinical pathogenic fungi Gramnegative bacteria E. coliand S. typhiwere revified on MacConkey Agar while
M. furfur and T. rubrum were revified on Potato Dextrose Agar (PDA). The bacteria were incubated for 24 h in 32oC while the
fungi were incubated in at 27oC for 24 h.
Fungal identification
Morphological approach
Fungus MT23 was recultured on MEA for 5 days with slide cultured method (Qiu
et al.2005). Sterilized MEA was cut with size 2 cm2 then transferred and put onto object
glass then covered by cover glass. After that, fungal mycelia were inoculated on each side of the MEA then incubated at room temperature (set at 23oC). After 5 days of incubation, object
and cover glasses were covered by fungal mycelia. MEA was separated from the glasses. Lactophenol cotton blue was dropped on the object and cover glasses then covered by its pairs. After that, the morphology of the fungi was observed under microscope.
Molecular approach
DNA of fungus MT23 was extracted using Chelex method from Turanet al.(2015) with particular modifications. Seven days old of mycelium were immersed in 100 µL ddH2O and 1000 µL of 0.5% saponin for 24 h in chilling temperature (4oC). Then it was separated by
centrifugator with 12000 rpm for 10 min. The supernatant was discarded while the template was added 100 µL of ddH2O followed by addition of 50 µL of 20% Chelex 100. Then it was heated in water bath at 80oC for 5 min.
After that, the mixture was homogenized using vortex for 10 s and reheated for 5 min. After reheated, the DNA in mixture were separated using centrifugation at 12000 rpm for 10 min. Then the supernatant were transferred to new microtube and stored at 20oC for 24h.
DNA of fungus MT23 was amplified based on Internal Transcribed Spacer (ITS) region. The amplification using primer 1 µL of ITS 1 (5’TCC GTA GGT GAA CCT GCG G3’) and 1 µL of ITS 4 (5’TCC TCC GCT TAT TGA TAT GC3’). The primers were mixed
with 12.5 µL of GoTaq Green Master Mix Promega, 0.5 µL of DNA extract and 10 µL of ddH2O. The PCR profile consisted of 3 min of preheat at 93oC for 3 min, 30 cycles for
denaturation at 95oC (1 min), annealing at
56.1oC (1 min) and extension at 72oC (1 min).
Final extension was done at 72oC for 7 min.
The quality of PCR product was checked using gel electrophoresis in agarose 1%. Then PCR product was sent to 1stBase Laboratories Sdn
Bhd, Malaysia for DNA sequencing. DNA sequence was analyzed to its homology using
Basic Local Alignment Search Tool
(www.ncbi.nlm.nih.gov). Phylogenetic tree was reconstructed using MEGA 7.0 software package (Tamuraet al.2011). Neighborjoining was applied as the statistical method with 1000 bootstrap replications.
Pigment extraction
Yellow pigment from fungus MT23 was extracted using solidliquid extraction (Manikkam et al. 2015). The mycelia were separated from media, then the media were cut into smaller size and dried using silica gel in desiccator for 48 h. After that, solvent optimization for pigment extraction was done by immersing the dried agar in methanol, ethyl acetate and nhexane with agitation using a shaker (118 rpm) for 24 h then filtered using filter paper (MachereyNagel 640d·Ø 1125 mm). The filtrate was evaporated using rotary evaporator (Eyela SB1100) set at 38oC.
The pigment yield percentage was calculated using following equation:
Determination of inhibition zone
Antibacterial activity
Antibacterial activity of the pigment was evaluated using agardisc diffusion method from Balouiriet al.(2016) with several modifications. Pigment extract was dissolved in methanol and diluted to five concentrations (50, 125, 250, 500 and 100 µg/mL).S. typhi
and E. coli strain MDR were grown on MacConkey Agar for 24 h (32oC) then diluted
in physiological saline solution into 0.5
McFarland. Then the bacterial solution was inoculated to MHA using sterile cotton swab. Each extract (10 µl) was injected onto sterilized paper disc then placed on the MHA and incubated at 32oC for 24 h. Amoxicillin +
Clavulanic acid (30 µg/disc) was used as positive control. The presence of clear zone indicated the antibacterial activity of the pigment extract.
Antifungal activity
This assay was done according to Bhalodia and Shukla (2011) and Balouiriet al.(2016) with modifications. Pigment extract was dissolved in methanol and diluted to five concentrations (50, 125, 250, 500 and 100 µg/mL).M. furfurandT. rubrumwere grown on PDA for 24 h at 27oC. Then the fungi were
inoculated using pour plate method. Each extract (10 µL) was injected onto sterilized paper disc then placed on the PDA and incubated at 27oC for 24 h. Nystatin was used
as positive control in this study. The presence of clear zone on PDA indicated the antifungal activity of the pigment extract.
Data analysis
The impact of solvents to pigment yields was calculated with One Way Analysis of Variance (ANOVA) with P<0.05 while antibacterial and antifungal activities were calculated using Two Way ANOVA with P<0.05. The data was analyzed using SPSS 5.0.
Results and Discussion
Fungal identification
Fungus MT23 is a marine sponge associated fungi which is isolated from sponge from Teluk Awur, Jepara, Central Java. Macroscopic characteristics of fungus MT23 were white colour mycelium with green radial growth ring, hyphae concentrated and produced yellow colour in the media. This fungus grown well in room temperature (23oC) and changed the colour of MEA from
slightly orange to yellow colour. Colour change was started at fourth day of cultivation and successfully changed the colour of whole media at day 14 (Figure 1). Physical conditions of environment including pH and temperature
were reported to give impact to the fungal pigment production. A fungus Penicillium purpurogenumGH2 produced highest yield of red pigment after 240 h (10 days) of cultivation with pH 5 at 24oC (Mendézet al.
2011). On the other hand, Velmurunganet al.
(2010) induced fungal pigment production using colour of lights. They figured out that the darker lights like red (780–622 nm) and blue (492–455) gave higher pigment productions compared to green (577–492 nm), yellow (597–577 nm) and white lights in 5 different fungi. This phenomenon could be explained by the postulating of the existance of photoreceptor in fungi. However, there was no clear explanation about the mechanisms of the light to induce pigment productions in fungi. Furthermore, Blumenstein et al.
(2005) suggested that a phytochrome type of system may be operative in particular fungi, in his case wasA. nidulans. The phytochrome suggested able to sense the red and farred light through photointerconversion between the two stable conformations.
Figure 2 shows the morphology of fungus MT23 under microscope. This fungus had hyaline and thin mycelium with septum. The conidiophore were branching with lageniform phialide with green conidia, smooth, eguttulate, and ellipsoidal to cylindrical form. According to its characteristics, fungus MT23 was judged as member of genus
Trichoderma(Atanasovaet al., 2010; Rahman
et al., 2011; Qin and Zhuang, 2016). To confirm the accuracy of morphological identification, we did identification through molecular approach.
Figure 1.a) 4 days old of mycelium, b) 4 days old of reverse side, c) 14 days old of reverse side.
I.J. Biotech.
region, internal transcribed spacer 2 region and the beginning of 28S rRNA gene region (Whiteet al., 1990; Larenaet al., 1999; Anderson
et al., 2003; Bellemainet al., 2010; Schochet al., 2012). Figure 3 shows the location which amplified by ITS primer.
Fungal DNA amplification was done by using polymerase chain reaction (PCR) the quality of PCR product was checked using gel electrophoresis. The result of visualization of fungal DNA quality is shown by Figure 4.
Figure 4 shows that the fungal DNA was successfully amplified in region between 500 to 1000 bp. This result is supported by the statement of Nilsson et al. (2009) who stated that the fungal DNA is normally obtainable in a single round of Sanger DNA sequencing in 650 bp region. PCR product was sequenced to get the sequence of fungal
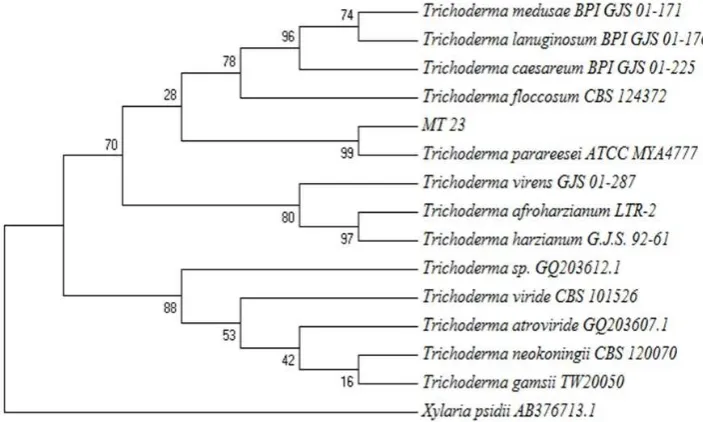
DNA, and then the sequence was compared to that of the database in gene bank using BLAS homology. According to fungal DNA comparison, fungus MT23 was 99% similar to Trichoderma parareesei ATCC MYA4777 (Houseknechtet al., 2011). Phylogenetic tree of fungus MT23 is shown by Figure 5.
Result of morphology observation was supposed fungus MT23 was member of genus Trichoderma and supported by the molecular identification which successfully identified fungus MT23 as T. parareesei.
Genus Trichoderma is well known as
terrestrial fungus and give advantages for agricultural sector (Rinu et al., 2013; Saravanakumaret al., 2013; El Komyet al., 2015; Hamedet al., 2015). The finding ofT. parareeseias marine sponge associated fungi showed that this fungus was a marine facultative (marine derived) fungi. Several species from Trichoderma such as T. longibrachiatum, T. harzianum, andT. atroviride
were reported to tolerate to the increasing of salinity (GalHemedet al.2011). Genus of
Trichodermawas also known to produced pigment and bioactive compounds (Rubeena
et al., 2013; Vacondioet al., 2015; Benkadaet al., 2016).
Pigment extraction
Fungus MT23 changed the colour of MEA to yellow because it produced extracellular pigment. Extracellular pigments production is indicated by colour changing of the environment where the fungi live. Previous research successfully identified black extracellular pigment which produced by endophytic fungi as melanin having photoprotector activity (Siberoet al., 2016). Fungus MT23 produced yellow pigment started at the fourth day of cultivation and Figure 2. a) conidiophore, b) mycelium, c) conidia.
Figure 3.Diagram of the rDNA region of fungi and ITS primer (Source: Bellemain et al., 2010).
Figure 4.Visualization of DNA band of fungus MT23.
ready to be harvested on day fourteenth. Pradeep and Pradeep (2013) studied the optimum cultivation condition for pigment production of fungusFusarium moniliforme. The fungus had highest pigment absorbance (AU 500nm) at temperature 28–30oC, pH 5.5,
glucose as carbon source, peptone as nitrogen source and cultivated for 8 days. Yellow pigment from fungus MT23 were extracted using organic solvents. Environmental conditions have important role in fungi growth. Pradeep and Pradeep (2013) studied the optimum cultivation condition for growth of fungusFusarium moniliforme. The fungus had highest biomass (g/L) at temperature 28oC, pH 5.5, glucose as carbon source,
peptone as nitrogen source, KH2PO4as source of salt and addition of methionine as additional amino acid. Stationary phase of this fungus was on eighth day of cultivation. In this study, we used Malt Extract Agar (MEA) from HiMedia M173 which contained malt extract and mycological peptone. Peptide and amino acid in peptone are easily metabolized by fungi which induced the production of fungal metabolites, including pigment (Calestinoet al., 2014). Da Costa Souza et al. (2016) used several media including malt extract to induced pigment production in filamentous fungi. As a result, pigment production in malt extract was higher than other medium. In addition,
nutrient content in media can regulate the expression of genes to activate the metabolic path way for pigments production (Pradeep
et al., 2013).
Organic solvents such as nhexane, ethyl acetate, chloroform, methanol and dichloromethane were usually used to extract natural pigments from microorganisms (Saravanan and Radhakrishnan, 2016; Weber
et al., 2016). In this research, nhexane, ethyl acetate and methanol were used to extract fungal pigment. The purpose of this step was to know the best solvent to obtain highest fungal pigment yield. The result of pigment extraction is shown by Figure 6 and Table 1. Table 1 shows that only methanol was able to extract fungal extracellular pigment from fungus MT23. Statistic analysis showed that solvents gave significant difference to the fungal pigment yield (P<0.05). The polarity
of pigment will influence the suitability of Figure 5.Phylogenetic tree of fungus MT23.
optimum solvent to extract the pigment. Several pigments such as astaxanthin, chlorophyll a, chlorophyll b and xanthophyll were able to be extracted with methanol (Sasidharanet al., 2013, Sumantaet al., 2014; Bhat and Marar 2015).
Antibacterial and antifungal activities Fungal pigments are known to produce antimicrobial activity. Geweely (2011) successfully investigated antimicrobial of several fungal pigments. The result showed that fungus Aspergillus nidulans, Fusarium moniliforme, Penicillium purpurogenum, and
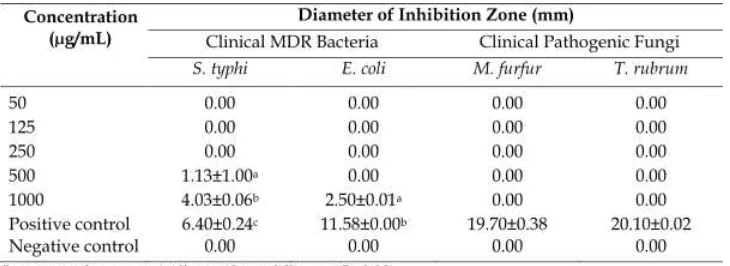
Phoma herbarum had weak to moderate antimicrobial activity against dermatophyte fungi, non dermatophyte fungi and several bacteria. Another research showed antibacterial activity of astaxanthin pigment from marine yeast (Ushakumari and Ramanujan, 2013). Fungal pigment from fungus MT23 was tested against several clinical pathogenic microorganisms. The result of antibacterial and antifungal is shown in Table 2.
According to Table 2, fungal pigment from MT23 showed weak antibacterial activity against clinical MDR bacteriaS. typhiandE. coli. The biggest antibacterial activity showed at the concentration of 1000 µg/mL against
S. typhi. The pigment didn’t show any antifungal activity against clinical pathogenic fungi. The
weak antimicrobial activity means that this fungal pigment is not potential as a source of antibiotic against clinical pathogenic and MDR microorganisms. Carotenoid is one of natural pigment which produced by fungi with yellow, orange to reddish colors. The production of this pigment is related to stress tolerance or with the synthesis of physiologically active by product (Avalos and Limόn, 2015). Carotenoid pigment is mostly reported has antimicrobial and antioxidant potential (Ernawita et al., 2016; Yoo et al., 2016). The antimicrobial mechanisms of this pigment could lead to the accumulation of lysozyme enzyme that digest bacterial cell walls (Abu Ghannamet al., 2013).
Conclusions
Sponge associatedfungus isolate MT23 was successfully identified using morphological and molecular approaches as
member of Trichoderma and had 99%
similarity to Trichoderma parareesei. The fungal pigment could be extracted only in methanol with yield 6,22±0,29%. The pigment showed weak antibacterial activity against
S. typhiandE. colistrain MDR, furthermore the pigment did not show any activity against clinical pathogenic fungi.
Acknowledgements
We thank Directorate of Research and Community Development, Ministry of Research, Technology and Higher Education for the research grant through Program Magister Menuju Doktor Bagi Sarjana Unggulan (PMDSU) scheme, funding in the year 2016, No: 299312/UN7.5.1/PG/2016.
Solvent
Methanol 6.26±0.29
nHexane Ethyl acetate
Fungal pigment yield (%)
0 0
Table 1.Fungal pigment yield with different solvents.
Siberoet al. I.J. Biotech.
References
AbuGhannam, N., and Rajauria, G. (2013). Antimicrobial activity of compounds isolated from algae. In Functional Ingredients from Algae for Foods and Nutraceuticals; Domínguez, H., Ed. Cambridge (UK): Woodhead Publishing.
Anderson, I.C., Campbell, C.D., Prosser, J.I. 2003. Potential bias of fungal 18S rDNA and internal transcribed spacer polymerase chain reaction primers for estimating fungal biodiversity in soil. Environ. Microbiol., 5(1), 3647. Atanasova, L., Jaklitsch. W.M., Komoń
Zelazowska, M., Kubicek, C.P., Druzhinina, I.S. 2010. Clona species
Trichoderma parareeseisp. nov. likely resembles the ancestor of the cellulase producerHypocrea jecorina/T. reesi. Appl. Environ. Microbiol., 76(21), 72597267.
Avalos, J., Limon, M.C. 2015. Biological roles of fungal carotenoids. Curr. Genet., 61(3), 309324.
Balouiri, M., Sadiki, M., Ibnsouda, S.K. 2016. Methods for in vitro evaluating antimicrobial activity: A review. J. Pharm. Anal., 6, 7179.
Bellemain, E., Carlsen, T., Brochmann, C., Coissac, E., Taberlet, P., Kauserud, H. 2010. ITS as an environmental DNA barcode for fungi: an in silico approach reveals potential PCR biases. BMC Microbiol., 10(89), 1–9.
Benkada, M.M., Pouchus, Y.F., Vérite, P., Pagniez, F., Caroff, N., Ruiz, N. 2016. Identification and biological activities of longchain peptaibols produced by a marinederived strain ofTrichoderma longibrachiatum. Chem. Biodiver., 13, 521–530.
Bhalodia, N.R., and Shukla, V.J. 2011. Antibacterial and antifungal activities from leaf extracts ofCassia fistulal.: An ethnomedicinal plant. J. Adv. Pharm. Technol., 2(2), 104109.
Bhat, M.R., and Marar, T. 2015. Media optimization, extraction and partial characterization of an orange pigment
fromSalinicoccussp. MKJ 997975. Int. J. Life. Sci. Biotech. Pharm. Res., 4(2), 8589.
Blumenstein, A., Vienken, K., Purschwitz, J., Tasler, R., Frankenberg, N., Fischer, R. 2005. The phytochrome FphA controls development in the filamentous fungusAspergillus nidulans. Curr. Biol., 15, 1833–1838.
Celestino, J., dos, R., de Carvalho, L.E., Lima, M. da P., Lima, A.M., Ogusku, M.M., de Souza, J.V.B. 2014. Bioprospecting of Amazon soil fungi with the potential for pigment production. Process Biochem., 49, 569–575.
de Costa Souza, P.N., Grigoletto, T.L.B., de Moraes, L.A.B., Abreu, L.M., Guimaraes, L.H.S., Santos, C., Galvao, L.R., Cardoso, P.G. 2016. Production and chemical characterization of pigments in filamentous fungi. Microbiol., 162, 1222. DelgadoVargas, F., Jiménez, A.R., Paredes Lόpez, O. 2000. Natural pigments: carotenoids, anthocyanins, and betalainscharacteristics, biosynthesis, processing, and stability. Crit. Rev. Food. Sci., 40(3), 173289.
Dong, Z., Saikumar, P., Weinberg, J.M., Venkatachalam, M.A. 1997. Inter nucleosomal DNA cleavage triggered by plasma membrane damage during necrotic cell death. Involvement of serine but not cysteine proteases. Am. J. Pathol., 151(5), 12051213.
El Komy, M.H., Saleh, A.A., Eranthodi, A., Molan, Y.Y. 2015. Characterization of novelTrichoderma asperellumisolates to selects effective biocontrol agents against tomatoFusariumwilt. Plant Pathol. J., 31(1), 5060.
Ernawita, Wahyuono, R.A., Hesse, J., Hipler, U.C., Elsner, P., Böhm, V. 2016. Carotenoids of indigenous citrus pecies from Aceh and its in vitro antioxidant, antidiabetic and antibacterial activities. Eur. Food. Res. Technol., 242(11), 1869 1881.
Trichodermaspp. as potential halotolerant agents of biological control for arid zone agriculture. App. Environ. Microbiol., 77(15), 51005109.
Geweely, N.S. 2012. Investigation of the optimum condition and antimicrobial activities of pigments from four potent pigmentproducing fungal species. J. Life Sci., 5, 697711.
Hamed, R.E., Awad, H.M., Ghazi, E.A., El Gamal, N.G., Shehata, H.S. 2015.
Trichoderma asperellum isolated from salinity soil using rice srtaw waste as biocontrol agent for cowpea plant pathogens. J. App. Pharm. Sci., 5(2), 9198. Houseknecht, J.L., Suh, S.O., Zhou J. 2011.
Trichoderma parareeseiATCC MYA4777 ITS region; from TYPE material. https://www.ncbi.nlm.nih.gov/nucleoti de/1051341416?report=genbank&log$=n uclalign&blast_rank=2&RID=5RZZ5WV G01R (Accessed on 22 Januari 2016). Ibrahim, D, Nazari, T.F., Kassim, J, Lim S.H.
2014. Prodigiosinan antibacterial red pigment produced bySerratia marcescens
IBRL USM 84 associated with a marine spongeXestospongia testudinaria. J. App. Pharm. Sci., 4(10), 001006.
Idraningrat, A.A.G., Smidt, H., Sipkema, D. 2016. Bioprospecting spongeassociated microbes for antimicrobial compounds. Mar. Drugs, 14(5), 87.
Larena, I., Salazar, O., González, V., Julián, M.C., Rubio, V. 1999. Design of a primer for ribosomal DNA internal transcribed spacer with enhanced specidy for Ascomycetes. J. Biotechnol., 75, 187194. Liu, L., Wang, C.L., Peng, W.Y., Yang, J., Lan, M.Q., Zhang, B., Li, J. B., Zhu, Y.Y., Li, C.Y. 2015. Direct DNA extraction method on an obligate parasitic fungus from infected plant tissue. Genet. Mol. Res., 14(4), 1854618551.
Magiorakos, A.P., Srinivasan, A., Carey, R.B., Carmeli, Y., Falagas, M.E., Giske, C.G., Harbarth, S., Hindler, J.F., Kahlmeter, G., OlssonLiljequist, B., Paterson, D.L., Rice, L.B., Stelling, J., Struelens, M.J., Vatopoulos, A., Weber, J.T., Monnet, D.L. 2012. Multidrugresistant,
extensively drugresistant and pandrug resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infec., 18(3), 268281. Manikkam, R., Venugopal, G., Ramasamy,
B., Kumar, V. 2015. Effect of critical medium components and culture conditions on antitubercular pigment production from novel Streptomyces
sp. D25 isolated from Thar desert, Rajasthan. J. App. Pharm. Sci., 5(06), 015019.
Méndez, A., Pérez, C., Montanez, J. C., Martinéz, G., Aguilar, C. N. 2011. Red pigment production by Penicillium purpurogenum GH2 is influenced by pH and temperature. J. Zhejiang Univ., 12(12), 961968.
Nilsson, R.H., Ryberg, M., Abarenkov, K., Sjökvist, E., Kristiansson, E. 2009. The ITS region as a target for characterization of fungal communities using emerging
sequencing technologies. FEMS
Microbiol. Lett., 296, 97101.
Norman, M., Bartczak, P., Zdarta, J., Ehrlich, H., Jesionowski. 2016. Anthocyanin dye conjugated withHippospongia communis
marine demosponge skeleton and its antiradical activity. Dyes Pigm., 134, 541552.
Pradeep, F.S. and Pradeep, B.V. 2013. Optimization of pigment and biomass production fromFusarium moniliforme
under submerged fermentation
conditions. Int. J. Pharm. Pharm. Sci., 5(3), 526535.
Pradeep, F.S., Begam, M.S., Palaniswamy, M., Pradeep, B.V. 2013. Influence of culture media on growth and pigment production by Fusarium moniliforme
KUMBF1201 isolated from paddy field soil. World Appl. Sci. J., 22, 70–77. Qin, W.T. and Zhuang, W.Y. 2016. Seven
woodinhabiting new species of the genusTrichoderma(Fungi, Ascomycota) in Viride clade. Sci. Rep., 6(27074), 114. Qiu W.Y., Yao, Y.F., Zhu, Y.Y., Zhang, Y.M., Zhou P., Jin, Y.Q., Zhang, B. 2005. Fungal Spectrum Identified by a New
Slide Culture and In Vitro Drug susceptibility Using Etest in Fungal Keratitis. Curr. Eye. Res., 30, 11131120. Radhakrishnan, M., Gopikrishnan, V., Vijayalakshmi, G., Kumar, V. 2016. In vitro antioxidant activity and antimicrobial activity against biofilm forming bacteria by the pigment from desert soilStreptomycessp D25. J. Appl. Pharm. Sci., 6(6), 148150.
Rahman, A., Begum, M.F., Rahman, M., Bari, M.A., Illas, G.N.M., Alam, M.F. 2011. Isolation and identification of
Trichoderma species from different habitats and their use for bioconversion of solid waste. Turk. J. Biol., 35, 183194. Rinu, K., Sati, P., Pandey, A. 2013.Trichoderma
gamsii (NFCCI 2177): A newly isolated endophytic, psychrotolerant, plant growth promoting, and antagonistic fungal strain. J. Basic. Microbiol., 54(5), 408417.
Rostami, H., Hamedi, H., Yolmeh, M. 2016. Some biological activities of pigments extracted from Micrococcus roseus
(PTCC 1411) and Rhodotorula glutinis
(PTCC 5257). Int. J. Immunopathol. Pharmacol., 29(4), 684695.
Rubeena, M, Neethu, K, Sajith, S, Sreedevi, S, Priji, P, Unni, K.N., Josh, M.K.S, Jisha, V.N., Pradeep, S, Benjamin, S. 2013. Lignocellulotic activities of novel strain ofTrichoderma harzianum. Adv. Biosci. Biotechnol., 3, 214221.
Rymbai, H, Sharma, R.R,, Srivastav, M. 2011. Biocolorants and its implications in health and food industrya review. Int. J. PharmTech. Res., 3(4), 22282244. Salem, N., Msaada, K., Elkahoui, S., Mangano,
G., Azaeiz, S., Slimen, I.B., Kefi, S., Pintore, G., Limam, F., Marzouk, B. 2014. Evaluation of antibacterial, antifungal, and antioxidant activities of Safflower natural dyes during flowering. BioMed. Res. Int., 2014, 110. Saravanakumar, K., Arasu, V.S., Kathiresan, K. 2013. Effect of Trichoderma on soil phosphate solubilization and growth improvement of Avicennia marina. Aquat. Bot., 104, 101105.
Saravanan, D., and Radhakrishnan, M. 2016. Antimicrobial activity of pigments produced by fungi from Western Ghats. J. Chem. Pharm. Res., 8(1), 634638. Sasidharan, P., Raja, R., Karthik, C., Sharma,
R., Arulselvi, P. I. 2013. Isolation and characterization of yellow pigmented producing Exiguobacterium sps. J. Biochem. Toxicol., 4(4), 632635. Schoch, C.L., Seifert, K.A., Huhndorf, S.,
Robert, V., Spouge, J.L., Levesque, C. A., Chen, W., Concortium, F.B. 2012. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for fungi. Proc. Natl. Acad. Sci., 109(16), 62416246.
Sibero, M.T., Tarman, K., Hanif, N. 2016. Characterization and photoprotector activity of endophytic fungal pigments from coastal plant sarang semut (Hydnophytum formicarum). J. Pengolah. Hasil. Perikan. Indones., 19(1), 18. Srilekha, V., Krishna, G., Srinivas, S.V.,
Charya, S.M.A. 2016. Isolation and screening of marine pigmented bacteria from Nellore Coast for antimicrobial studies. J. Cell. Tissue. Res., 16(1), 5413 5419.
Sumanta, N, Haque, C.I., Nishika, J, Suprakash, R. 2014. Spectrophotometric
analysis of chlorophylls and
carotenoids from commonly grown fern species by using various extracting solvents. Res. J. Chem. Sci., 4(9), 6369. Suresh, M., Renugadevi, B., Brammavidhya, S., Iyapparaj, P., Anantharaman, P. 2015. Antibacterial ctivity of red pigment produced byHalolactibacillus alkaliphilus
MSRD1—an isolate from seaweed. Appl. Biochem. Biotechnol., 176(1), 185 195.
Susilowati, R., Sabdono, A., Widowati, I. 2015. Isolation and characterization of bacteria associated with brown algae
Sargassum spp. from Panjang Island and their antibacterial activity. Procedia Environ. Sci., 23, 240246.
analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol., 28(10), 27312739.
Turan, C., Nanni, I.M., Brunelli, A., Collina, M. 2015. New rapid DNA extraction method with Chelex from Venturia ineaqualisspores. J. Microbiol. Methods, 115, 139143.
Ushakumari, U.N.U., and Ramanujan, R. 2013. Isolation of astaxanthin from marine yeast and study of its pharmacological activity. Int. Curr. Pharm. J., 2(3), 6769. Vacondio, B., Birolli, W., Ferreira, I., Porto, A.L.M. 2015. Biodegradation of pentachlorophenol by marinederived fungusTrichoderma harzianumCBMAI 1677 isolated from ascidianDidemnun ligulum. Biocatal. Agric. Biotechnol., 4(2), 266275.
Velmurungan, P., Lee, Y.H., Venil, C.K., Laksmanaperumalsamy, P., Chae, J.C., Oh, B.T. 2010. Effect of light on growth, intracellular and extracellular pigment production by five pigmentproducing filamentous fungi in synthetic medium. J. Biosci. Bioeng., 109(4), 345350. Venil, C.K., Zakaria, Z.A., Ahmad, W.A. 2013.
Bacterial pigments and their
application. Process Biochem., 48, 1065 1079.
Walsh, P.S., Metzger, D.A., Higuchi, R. 2013. Chelex 100 as a medium for simple extraction of DNA for PCRbased typing from forensic material. BioTechniques, 54(3), 134139.
Weber, G.L., Boonloed, A, Naas, K.M., Koesdjojo, M.T., Remcho, V.T., Robinson, S.C. 2016. A method to stimulate production of extracellular pigments from wooddegrading fungi using a water carrier. Curr. Res. Environ. Appl. Mycol., 6(3), 218230. White, T.J., Bruns, T., Lee, S., Talyor, J. (1990)
Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis, M.A., Gelfand, D.H., Sninisky, J.J. and White, T. J. Eds. PCR protocols: a guide to methods and applications.
San Diego (US): Academic Press. pp. 315322.
Wu, B., Oesker, V., Wiese, J., Schmaljohann, R., Imhoff, J. F. 2014. Two new antibiotic pyridones produced by a marine fungus
Trichoderma sp. strain MF106. Mar. Drugs, 12, 12081219.
Yolmeh, M., Hamedi, H., Khomeiri, M. 2016. Antimicrobial activity of pigments extracted from Rhodotorula glutinis
against some bacteria and fungi. Zahedan J. Res. Med. Sci., 18(12), 15. Yoo, A.Y., Alnaeeli, M., Park, J.K. 2016.
Production control and characterization of antibacterial carotenoids from the yeastRhodotorula mucilaginosaAY01. Process Biochem., 51(4), 463473.