Purification, Characterization, and cDNA Cloning of a Novel Lectin
from the Green Alga,
Codium barbatum
Danar P
RASEPTIANGGA, Makoto H
IRAYAMA, and Kanji H
ORIyFood Science and Biofunctions Division, Graduate School of Biosphere Science, Hiroshima University,
1-4-4 Kagamiyama, Higashi-Hiroshima 739-8528, Japan
Received December 8, 2011; Accepted December 28, 2011; Online Publication, April 7, 2012 [doi:10.1271/bbb.110944]
A novel lectin (CBA) was isolated from the green
alga,
Codium barbatum
, by conventional
chromato-graphic methods. The hemagglutination-inhibition
pro-file with sugars and glycoproteins indicated that CBA
had preferential affinity for complex type
N
-glycans but
not for monosaccharides, unlike the other known
Codium
lectins
specific
for
N
-acetylgalactosamine.
CBA consisted of an SS-linked homodimer of a
9257-Da polypeptide containing seven cysteine residues, all of
which were involved in disulfide linkages. The cDNA of
the CBA subunit coded a polypeptide (105 amino acids)
including the signal peptide of 17 residues. The
calcu-lated molecular mass from the deduced sequence was
9705 Da, implying that the four C-terminal amino acids
of the CBA proprotein subunit were post-translationally
truncated to afford the mature subunit (84 amino acids).
No significantly similar sequences were found during an
in silico
search, indicating CBA to be a novel protein.
CBA is the first
Codium
lectin whose primary structure
has been elucidated.
Key words:
Codium barbatum
; lectin;
carbohydrate-binding specificity; cDNA cloning; primary
structure
Lectins, or carbohydrate-binding proteins, are widely
distributed in nature and play some important roles as
recognition molecules in cell-cell or cell-matrix
inter-actions.
1)However, the physiological functions of most
lectins remain to be investigated in more detail,
especially for those of plant origin.
1,2)The
carbohy-drate-binding specificity and molecular structure of
lectins are diverse and generally depend on the
organ-isms from which they originate, although lectins have
been classified into several families based on the
amino acid sequences of their carbohydrate-recognition
domains, some of which are also evolutionarily
con-served.
3,4)The diversity of lectins in their
carbohydrate-binding specificity enables them to be used as
conven-ient tools to decode the carbohydrate structures on a cell
surface and in a body fluid. Recent studies have been
directed to the nutritional aspects of lectins, because
many edible plants, including cereals, vegetables, and
fruits, contain lectin proteins, and some plant lectins
have been reported to affect the transport of other food
gradients in an intestinal tract model.
1,5)Lectins have been isolated and characterized from
macroalgae and microalgae. These ‘algae-specific’
lectins have shown novel carbohydrate-binding
specific-ity and molecular structures, including those specific
for high-mannose
N
-glycans
6–14)and core (
1,6)
fuco-sylated
N
-glycans.
15,16)They have also shown some
interesting biological activities, including
anticarcino-genic
17)and antiviral effects.
6–9,11–13)Their antiviral
activities are especially remarkable because they inhibit
in vitro
infection by the immunodeficiency virus
(HIV-1) and influenza virus with a half-maximal effective
concentration (EC
50) in the low nanomolar to picomolar
range through their binding with viral envelope
glyco-proteins.
18)Many algal lectins, especially from red
algae, share some common characteristics of low
mo-lecular weight, monomeric form, thermostability, and
divalent cation-independent hemagglutination, and
hav-ing affinity only for glycoproteins and not for
mono-saccharides. These properties of algal lectins are
dissimilar to most land plant lectins that have affinity
for monosaccharides and consist of oligomeric forms.
2)Monosaccharide-binding lectins have recently been
reported from several species of green algae belonging to
the genera
Enteromorpha
,
19)Ulva
,
20,21)and
Codium
.
22–24)Among these, the lectins from the
Codium
,
C. fragile
ssp.
tomentosoides
,
22)C. fragile
ssp.
atlanticum
,
22)C.
tomentosum
,
23)and
C. giraffa
24)are commonly specific
for
N
-acetylgalactosamine and/or
N
-acetylglucosamine
and consist of oligomeric forms, except for a monomeric
lectin from
C. giraffa
. The resemblance of the lectin
properties may be derived from the close evolutionary
distance between the green algae of the genus
Codium
and the land plant, although there is no information
on the primary structures of
Codium
lectins. The lectin
from
C. fragile
ssp.
tomentosoides
has shown
prefer-ential affinity for the Forssman antigen sugar chain
that is a marker of cancer.
25)Thus, the lectins of the
genus
Codium
are interesting targets for their application
as biochemical and clinical reagents, as well as to
provide insight into the molecular evolution of lectins in
the plant kingdom. The characterization of
Codium
lectins may also be significant in evaluating their
y To whom correspondence should be addressed. Tel/Fax: +81-82-424-7931; E-mail: [email protected]
Abbreviations: CBA,Codium barbatumlectin; BLAST, basic local alignment search tool; BSM, bovine submaxillary mucin; GNA,Galanthus
nivalislectin; DTT, dithiothreitol; ESI-MS, electrospray ionization-mass spectrometry; HIV, human immunodeficiency virus; Lys-C,
nutritional aspects, because
Codium
has been reported
as edible.
26)Our recent screening of lectins in marine algae
showed the extract from
C. barbatum
to have strong
hemagglutination activity. However, this activity was
not inhibited by any of the monosaccharides examined,
including
N
-acetylgalactosamine,
unlike
the
other
known
Codium
lectins. This present study deals with a
novel lectin isolated from
C. barbatum
.
Materials and Methods
Materials.TheC. barbatumgreen alga was collected from
Satsuma-Iwojima Island of Kagoshima in Japan, and was kept at30C until
being used. A portion of the C. barbatum tissue collected from Tanegashima Island of Kagoshima in Japan was also stored at30C
in RNAlater (Invitrogen) for subsequent RNA extraction. TSKgel Phenyl-5PW and TSKgel ODS 80TM columns were purchased from Tosoh Corporation (Tokyo, Japan), and the YMC Protein-RP column from YMC (Kyoto, Japan). The silica gel-immobilized phosphorylcho-line (PC 300S (N)) column was generously provided by Dr. K. Muramoto (Tohoku University, Japan). D-Glucose, D-galactose, N-acetyl-D-galactosamine, transferrin, fetuin, porcine thyroglobulin (PTG), bovine submaxillary mucin (BSM), and porcine stomach mucin (PSM) were purchased from Sigma-Aldrich Co. (MO, USA). D-Mannose,L-fucose,N-acetyl-D-glucosamine,N-acetylneuraminic acid, lactose, and yeast mannan were purchased from Nacalai Tesque Co. (Kyoto, Japan).D-Xylose andL-rhamnose were purchased from Wako Chemical Co. (Osaka, Japan). The desialylated (asialo-) derivatives of glycoproteins were prepared by hydrolyzing the parent sialoglycopro-tein with 0.05MHCl for 1 h at 80C, with subsequent dialysis overnight
against saline.27)A marker kit for molecular weight determination by SDS–PAGE was purchased from Tefco (Tokyo, Japan). As reference proteins in gel filtration, bovine serum albumin (67 kDa), chymotrypsi-nogen (25 kDa), and ribonuclease A (13.7 kDa) were obtained from Sigma-Aldrich Co., and ovalbumin (44 kDa) from Wako Chemical Co. Lysylendopeptidase (Lys-C) was purchased from Sigma-Aldrich Co., all other chemicals used in this study being of the highest purity available.
Purification of theC. barbatumlectin.A frozen sample (500 g) of
C. barbatumwas thawed, cut into small pieces, and ground in liquid
nitrogen into powder. The powdered alga was stirred overnight at 4C
with 500 mL of a 20 mMTris–HCl buffer (pH 7.5) containing 150 mM NaCl (TBS). The mixture was centrifuged at 13,500 g for 30 min, and to the supernatant, solid ammonium sulfate was added to attain a final concentration with 75% saturation. The mixture was kept overnight at 4C and centrifuged for 30 min at 13
;500g. The precipitate was dissolved and thoroughly dialyzed against TBS. The internal fraction was further centrifuged for 30 min at10;000g, and the supernatant was recovered as a salting-out fraction. This salting-out fraction was dialyzed against a 20 mMTris–HCl buffer at pH 7.5 (TB) containing 1Mammonium sulfate. Five mL each of the dialyzate was applied to the TSKgel Phenyl-5PW column (7:575mm) that had been
equilibrated with TB containing 1Mammonium sulfate. The column was washed for 20 min with the starting buffer and then eluted for 40 min with a linear gradient of ammonium sulfate concentration (1.0– 0M) in TB and then with TB alone for 30 min. The flow rate was 0.5 mL/min. Fractions of 2 mL were collected and measured for their absorbance at 280 nm and for hemagglutination activity. The active fractions showing hemagglutination activity were pooled, dialyzed against TBS, and subjected to gel filtration in the PC 300S (N) column (7:8300mm) that had been equilibrated with a 50 mMphosphate buffer at pH 7.0 containing 0.15M NaCl (PBS), according to the method reported by Watanabeet al.28)The column was eluted at a flow rate of 0.5 mL/min with PBS, and fractions of 2 mL were collected and measured for their absorbance at 280 nm and for hemagglutination activity.
Hemagglutination assay.Hemagglutination activity was determined
in a 96-well microtiter V-plate, using a 2% suspension (v/v) of trypsin-treated rabbit erythrocytes (TRBC).29) Serially two-fold dilutions
(25mL each) of a test solution were first prepared in saline, and to each well, 25mL of TRBC was added. The plate was gently shaken and allowed to stand at room temperature for 1 h. Hemagglutination was macroscopically observed and judged as positive when more than 50% of TRBC in the well had agglutinated. The hemagglutination activity is expressed as a titer, the reciprocal of the highest two-fold dilution exhibiting positive hemagglutination.
TRBC was prepared from rabbit blood purchased from the Laboratory of Animal Experiments (Hiroshima, Japan). The blood cells were washed three times with 50 volumes of saline and suspended in saline to give a 2% (v/v) suspension of native erythrocytes. A tenth volume of 0.5% (w/v) trypsin in saline was added to a 2% suspension of native erythrocytes, and the mixture was incubated for 60 min at 37C. The incubated erythrocytes were washed four times with saline,
and a 2% suspension (v/v) of TRBC was prepared in saline.
Determination of the protein contents.The protein contents were
determined by the method of Lowry et al.,30) using bovine serum albumin as a standard.
Effects of pH, temperature, and divalent cations on the
hemag-glutination activity. The effects of pH, temperature, and divalent
cations on the hemagglutination activity were next examined. To determine the effect of pH, 500mL each of a test solution was dialyzed overnight against 100 mL of a 50 mM buffer of various pH values between 3 and 10 at 4C. The pH-treated solution was further dialyzed
against PBS (pH 7.0) and subjected to the hemagglutination assay. The buffers used were glycine-HCl for pH 3.0, acetate for pH 4.0 and 5.0, phosphate for pH 6.0 and 7.0, Tris–HCl for pH 8.0, and carbonate for pH 9.0 and 10.0. To determine the effect of temperature, 500mL each of a test solution was incubated for 30 min at various temperatures from 30C to 100C and then subjected to the hemagglutination assay
after cooling. To determine the effect of divalent cations, 500mL of a test solution was dialyzed overnight at 4C against 50 mMEDTA in
PBS, and the internal fraction was measured for its hemagglutination activity. Additionally, to this internal fraction, an equal volume of 20 mMCaCl2or 20 mMMgCl2in saline was added, the mixture being kept for 2 h at room temperature and then measured for its hemagglutination activity.
Hemagglutination-inhibition assay.The inhibitory effects of sugars
and glycoproteins on the hemagglutination of the lectin solution were next examined. Serially two-fold dilutions (25mL each) of a sugar or a glycoprotein were first prepared in saline in a microtiter V-plate. To each well, an equal volume of the lectin solution with a hemaggluti-nation titer of 4 was added, and the plate was gently shaken and allowed to stand at room temperature for 1 h. Finally, 25mL of TRBC was added to each well, and the plate was gently shaken and allowed to stand for another 1 h. The inhibition of hemagglutination was macro-scopically observed, the inhibition activity being expressed as the minimum inhibitory concentration (mM or mg/mL), the lowest concentration of a sugar or glycoprotein at which complete inhibition of hemagglutination was achieved.
The sugars and glycoproteins used were D-glucose, D-mannose, D-galactose, N-acetyl-D-glucosamine, N-acetyl-D-galactosamine, L-fucose, N-acetylneuraminic acid, D-xylose, L-rhamnose, and lactose as sugars, and transferrin, asialo-transferrin, fetuin, asialo-fetuin, yeast mannan, PTG, asialo-PTG, BSM, asialo-BSM, and PSM as glycoproteins.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS–
PAGE).SDS–PAGE was performed by using 15% gel.31)The sample
was boiled at 100C for 5 min with or without 2% (v/v)
2-mercaptoethanol. The gel was stained with Commasie brilliant blue R-250 after electrophoresis.
Preparation ofS-pyridylethylated (PE-) CBA. S-Pyridylethylation
lectin solution, and the mixture incubated for another 2 h. The resulting PE-CBA, reduced andS-pyridylethylated (rPE)-CBA or non-reduced
and S-pyridylethylated (nrPE)-CBA was purified by reverse-phase
HPLC in the YMC Protein-RP column (6:0250mm), eluting with a
linear gradient between 10% and 70% acetonitrile in 0.1% TFA. The eluate was monitored by its absorption at 280 nm, and the peak containing PE-CBA was manually collected.
Enzymatic cleavage and separation of the peptides. rPE-CBA
(100mg) was digested at 37C for 18 h with lysylendopeptidase-C
(Lys-C) at an enzyme/substrate ratio of 1/100 (w/w) in 100 mMTB (pH 8.5) containing 4Murea. The peptide fragments were separated by reverse-phase HPLC in the YMC Protein-RP column (6:0250mm),
eluting with a linear gradient between 2% and 70% acetonitrile in 0.1% TFA. The eluate was monitored by its absorption at 220 nm, and the peaks were manually collected.
Molecular mass determination of the proteins and peptides.The
molecular masses of intact CBA, nrPE-CBA, rPE-CBA, and the peptides generated by enzymic digestion were determined by electro-spray ionization-mass spectrometry (ESI-MS), using LTQ Orbitrap XL (Thermo Fisher Scientific, MA, USA) and LCQ instruments (Finnigan, Bremen, Germany).
Determination of the N-terminal amino acid sequences.The
N-terminal amino acid sequences of rPE-CBA and its peptide fragments were determined by using a Procise 492 HT protein sequencing system (Applied Biosystems, CA, USA).
Rapid amplification of the 30and 50cDNA ends (30and 50RACE) of
CBA.Total RNA ofC. barbatumwas extracted from the RNAlater-treated algal tissues by using the Plant RNA Isolation reagent (Invitrogen). Full-length cDNAs were synthesized from 5mg of total RNA by using a GeneRacer kit (Invitrogen) according to the manufacturer’s instructions.
The first PCR for 30RACE was initiated by adding 0.2mL each of
a 10-fold dilution of the synthesized cDNA as a template to 8 aliquots of a 9.8-mL solution containing 1mL of a 10Blend Taq buffer (Toyobo, Osaka, Japan), 2 nmol each of dNTP, 6 pmol of the GeneRacer 30primer, 50 pmol of the degenerated CBA d F1 primer
(designed according to the N-terminal amino acid sequence of rPE-CBA (Table 1)), and 0.25 units of Blend Taq DNA polymerase (Toyobo). The reaction was performed with a T Gradient Thermo-cycler (Biometra, Go¨ttingen, Germany) under the following condi-tions: denaturation at 94C for 5 min, followed by 35 cycles consisting
of denaturation at 94C for 30 s, annealing at a gradient temperature of
50C to 64C (2C increments) for 30 s and extension at 72C for
90 s, and the final extension step at 72C for 5 min. The PCR products
in 8 aliquots were pooled, diluted 100-fold, and used as a template for nested PCR. Nested PCR was performed by the same method, except that 0.2mL of the dilution was used as a template, and 2 pmol of the GeneRacer 30nested primer and 50 pmol of the degenerated CBA d F2
primer (designed according to the sequence of the peptide fragment generated by Lys-C digestion) as a primer pair (Table 1), and an annealing temperature of 54C were used. The nested PCR products
were subcloned into the pGEM-T Easy vector (Promega, WI, USA)
and transformed intoEscherichia coliDH5competent cells. Plasmids from the transformants were purified with a HiYield Plasmid Mini kit (RBC Bioscience, New Taipei City, Taiwan) according to the manufacturer’s instructions. DNA sequencing was performed by using BigDye Terminator Cycle Sequencing kit ver. 3.1 with an ABI 3130xl genetic analyzer (Applied Biosystems).
50RACE was performed in a similar way to 30RACE just described,
except that the primer pair of GeneRacer 50 and CBA R1 designed
from the sequence obtained by 30RACE (Table 1) were used. The PCR
reaction was performed by using a high-fidelity DNA polymerase KOD Plus Neo kit (Toyobo) as follows: denaturation at 94C for
2 min, followed by 35 cycles consisting of denaturation at 98C for
10 s, annealing at gradient temperatures of 50C to 64C (2C
increments) for 30 s and then extension at 68C for 30 s, and the final
extension step at 68C for 5 min. Nested PCR was then performed by
the same method, except for using a 100-fold dilution of the first PCR products as a template, and the GeneRacer 50nested primer and CBA
R2 as the primer pair (Table 1). The PCR products were treated with a 10A-attachment mix (Toyobo), before subcloning into the pGEM-T Easy vector (Promega) and transforming intoE. coliDH5competent cells. Plasmid purification and DNA sequencing were performed as already described.
To verify the accuracy of the CBA sequences obtained by the 50and
30RACE operations, full-length cDNA of CBA was further amplified
by using KOD Plus Neo (Toyobo) and a primer pair of CBA 50end F
and CBA 30end R (Table 1) which were designed from the 50and 30
terminal sequences of CBA cDNA obtained by 50 and 30RACE.
Subcloning and DNA sequencing were then performed as already described.
Sequence data processing. Similarity searches for CBA were
performed by using the basic local alignment search tool (BLAST) with algorithms of protein-protein BLAST (BLASTP) and position-specific iterated (PSI) BLAST. The signal peptide region was predicted with SignalP 3.0.32)
Nucleotide sequence accession number.The sequence data for CBA
have been submitted to the DDBJ/EMBL/GenBank databases under accession no. AB675415.
Results
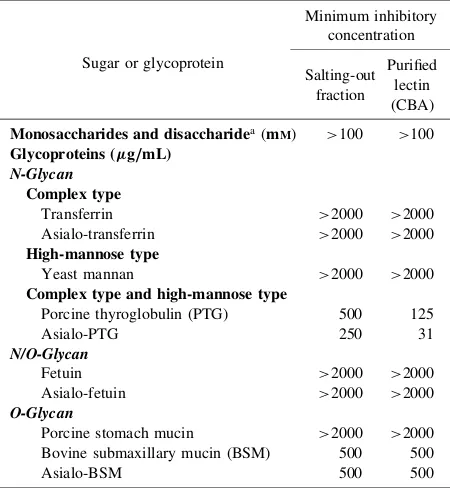
Purification of the
C. barbatum
lectin
The lectin from
C. barbatum
was extracted with TBS
and efficiently recovered as a precipitate with
75%-saturated ammonium sulfate (the salting-out fraction).
Hydrophobic chromatography in the TSKgel
Phenyl-5PW column for the salting-out fraction gave a single
peak of hemagglutination activity which was eluted as a
shoulder peak of protein (Fig. 1A). The active fractions
were pooled, dialyzed, and further purified by gel
filtration in the PC-300S (N) column to afford a single
peak (Fig. 1B). The purified lectin thus obtained gave a
single protein peak by reverse-phase HPLC in the YMC
Table 1. Primer Sequences Used in the cDNA Cloning of CBA
Primer Sequence (from 50to 30) Position
CBA d F1 ACICAYGGIATHAARAAYGAb
101–120
CBA d F2 AAYGAYTGYGGIGTICCIGTb 116–135
CBA R1 TCCAAGCAGCATACGAACAC 198–217
CBA R2 TCATCAGTCCCAGTCCAACA 124–143
CBA 50end F AATATAAATTAAACGTAAGGTATCGC 1–26
CBA 30end R AGTTCTGCATATGGATGTAC 457–476
GeneRacer 30primera
GCTGTCAACGATACGCTACGTAACG —
GeneRacer 30nested primera
CGCTACGTAACGGCATGACAGTG —
GeneRacer 50primera
CGACTGGAGCACGAGGACACTGA —
GeneRacer50nested primera
GGACACTGACATGGACTGAAGGAGTA —
a
These primers were inferred from the GeneRacer Kit (Invitrogen).
b
Protein-RP column (data not shown). The purified lectin,
named CBA, respectively gave a single protein band of
about 9 kDa and 18 kDa by reducing and non-reducing
SDS–PAGE (Fig. 1C), suggesting that CBA was the
SS-linked dimeric protein of a 9-kDa subunit. The relative
molecular mass of intact CBA was estimated to be about
18 kDa by gel filtration in the PC300S (N) column (data
not shown). The yield of CBA was 2.1 mg from 500 g
(wet weight) of the frozen alga. The result of purification
is summarized in Table 2.
Effects of pH, temperature, and divalent cations on
the hemagglutination activity of CBA
The hemagglutination activity of CBA was stable
under a wide range of pH values from 3 to 10 and was
not changed by incubating at 70
C for 30 min, although
it was inactivated when the incubation temperature
exceeded 80
C. The activity was not affected by either
the presence of EDTA or the addition of such divalent
cations as Ca
2þand Mg
2þ(data not shown).
Carbohydrate-binding specificity of CBA
The carbohydrate-binding specificity of CBA was
evaluated by a hemagglutination-inhibition assay with
a series of sugars and glycoproteins (Table 3). The
hemagglutination activity of CBA was not inhibited by
any of the monosaccharides and one disaccharide
examined. However, it was inhibited by such
glycopro-teins as PTG, asialo-PTG, BSM and asialo-BSM,
although not by the other glycoproteins examined,
including yeast mannan (Table 3). The strongest
inhib-ition was observed with asialo-PTG. The
hapten-inhib-ition profile of the purified lectin was almost the same as
that of the crude lectin (the salting-out fraction) (Table 3).
Molecular mass of CBA
The molecular mass of intact CBA was determined
to be 18500 Da by ESI-MS (data not shown). This is
comparable to the relative molecular mass (about
18 kDa) estimated by both non-reducing SDS–PAGE
(Fig. 1C) and gel filtration in the PC 300S (N) column.
The molecular masses of nrPE-CBA and rPE-CBA were
20.1 14.3 6.5
18.0 9.0
C 1 2 3 (kDa)
1.0
0.10
Absorbance at 280 nm Hemagglutination titer
Hemagglutination titer
Fraction
B
A
0 5 10 15 20 25
0 5 10 15
0.08
0.06
0.04
0.02
0 1
100 150
0 50
Absorbance at 280 nm
Fraction
(NH
4
)2
SO
4
(
M
[image:4.595.58.280.64.329.2])
Fig. 1. Purification of theC. barbatumLectin.
[image:4.595.307.537.75.187.2]A, Hydrophobic chromatography in a TSKgel Phenyl-5PW column of the precipitate obtained by salting-out with 75%-saturated ammonium sulfate. Fractions of 2 mL were collected and measured for their absorbance at 280 nm (filled circles) and for their hemagglutination activity (unfilled circles). The active fractions denoted by a bar were pooled. B, Gel filtration on a PC 300S (N) column of the active peak obtained by hydrophobic chromatogra-phy. Fractions of 2 mL were collected and measured for their absorbance at 280 nm (filled circles) and for their hemagglutination activity (unfilled circles). The active fraction denoted by a bar was recovered as the purified lectin (CBA). C, SDS–PAGE of the purified lectin fromC. barbatum(CBA): lane 1, molecular weight marker; lane 2, CBA with 2-mercaptoethanol; lane 3, CBA without 2-mercaptoethanol.
Table 2. Purification Procedure for the Lectin fromC. barbatum
Purification procedure Protein
a THAb MACc
(mg) (yield, %) (mg/mL)
Extraction 450.4 179200 2.51
(100.0) Ammonium sulfate-precipitation 184.6 323584 0.57
(75%-(NH4)2SO4) (180.6) Hydrophobic chromatography 8.0 10240 0.78
(5.7)
Gel filtration 2.1 3328 0.63
(1.9)
a
Protein contents were measured by the Folin-Lowry method (1951).30)
bTHA, total hemagglutination titer (volumehemagglutination titer, the
reciprocal of the highest two-fold dilution exhibiting positive hemaggluti-nation).
cMAC, minimum agglutination concentration, the protein concentration of
the highest dilution exhibiting positive hemagglutination.
Table 3. Hemagglutination-Inhibition Assay of the Salting-Out Fraction and a Purified Lectin (CBA) from C. barbatum with Monosaccharides, a Disaccharide, and Glycoproteins
Sugar or glycoprotein
Minimum inhibitory concentration
Salting-out Purified fraction lectin
(CBA)
Monosaccharides and disaccharidea
(mM) >100 >100
Glycoproteins (g/mL) N-Glycan
Complex type
Transferrin >2000 >2000
Asialo-transferrin >2000 >2000
High-mannose type
Yeast mannan >2000 >2000
Complex type and high-mannose type
Porcine thyroglobulin (PTG) 500 125
Asialo-PTG 250 31
N/O-Glycan
Fetuin >2000 >2000
Asialo-fetuin >2000 >2000
O-Glycan
Porcine stomach mucin >2000 >2000 Bovine submaxillary mucin (BSM) 500 500
Asialo-BSM 500 500
Values indicate the lowest concentration of sugar (mM) and glycoprotein (mg/mL) to completely inhibit the hemagglutination activity of a titer, 4. a
TheD-glucose,D-mannose,D-galactose,N-acetyl-D-glucosamine,N
[image:4.595.310.535.296.541.2]respectively determined to be 18500 Da and 9992 Da by
ESI-MS. These results indicate that CBA had no free
sulfhydryl groups, the mass of an SS-linked subunit
polypeptide (half size) being 9257 Da with reduced
sulfhydryl groups. The differential mass (735 Da)
be-tween rPE-CBA (9992 Da) and a CBA subunit
polypep-tide (9257 Da) corresponds to the total mass of seven
S
-pyridylethyl groups (105 Da
7), indicating that
CBA contained seven cysteine residues per subunit, all
of which were involved in disulfide linkages.
Primary structure of CBA
The amino acid sequence of CBA was principally
determined by cDNA cloning (Fig. 3A). The N-terminal
amino acid sequences of rPE-CBA and its peptide
fragments generated by digestion with Lys-C were first
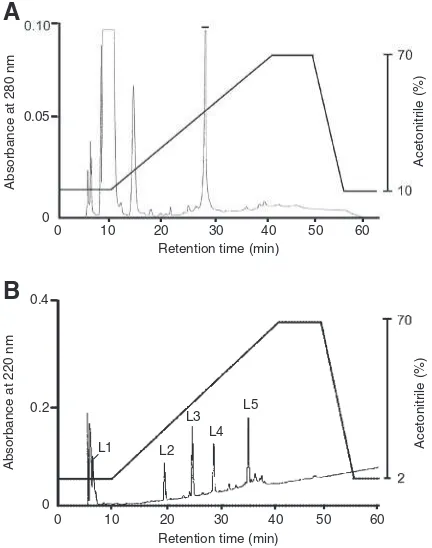
determined before cDNA cloning. Figure 2 shows the
elution pattern of rPE-CBA and its digested products by
reverse-phase HPLC in the YMC Protein-RP column.
The digested products gave several major peaks for
peptide fragments (L1–L5) in the HPLC data. The
peptide fragments thus obtained, as well as rPE-CBA,
were next sequenced for their N-terminal amino acids by
Edman degradation, except for L1, as shown in Fig. 3B.
The molecular masses of the peptide fragments
deter-mined by ESI-MS coincided with the calculated values
from the sequences, confirming the validity of the
sequencing operation (Fig. 3B). Although a potential
N
-glycosylation site (Asn-Gly-Ser) was found in the
sequence for the L5 fragment, it was not glycosylated
because Asn was identified at the position by Edman
degradation and the determined molecular mass of L5
coincided with the calculated value from its sequence
(Fig. 3B). In addition, the ESI-MS analysis shows that
L1 contained the two peptide fragments of ITHGIK
(667.4 Da) and VSTSYGSSK (914.4 Da), although their
N-terminal sequences were not analyzed.
cDNA cloning for CBA was performed by 3
0and
5
0RACE as described in the Materials and Methods
section to elucidate the complete amino acid sequence of
CBA. Full-length cDNA of the CBA subunit consisted
of 476 bp containing 46 bp of a 5
0-untranslated region
(UTR), 112 bp of 3
0UTR, and 318 bp of an open reading
frame (ORF) (Fig. 3A). ORF coded a polypeptide of 105
amino acids, including the signal peptide of 17 residues.
The N-terminal amino acid sequences of rPE-CBA and
the peptide fragments, which had been determined by
Edman degradation, were found in the sequence
de-duced from CBA cDNA (Fig. 3A), except that serine
was deduced from cloning at position 2, whereas
threonine was determined by Edman degradation. The
calculated molecular mass of the CBA subunit (88
amino acids) with serine at position 2 was 9705 Da. The
molecular mass of the CBA subunit substituted with
threonine at position 2 was therefore estimated to be
9719 Da which differs from the mass (9257 Da)
deter-mined by the ESI-MS analyses of intact CBA,
nrPE-CBA, and rPE-CBA. This difference indicates that the
A
B
Fig. 3. Nucleotide and Amino Acid Sequences of CBA.
A, The numbers represent the positions of nucleotides and amino acids. The underlined regions represent the signal peptide predicted with SignalP 3.0. The stop codon TGA is shown by an asterisk. B, The partial amino acid sequences determined by Edman degradation of rPE-CBA and its peptide fragments (represented by solid lines) are compared with the complete amino acid sequence deduced from CBA cDNA. Numbers above the sequences represent the position of the amino acids. Numeric values below the lines indicate the molecular masses (Da) of peptides determined by ESI-MS, and values in parentheses indicate those calculated from the sequences. These values are shown together with the S-pyridylethy-lated ones. The two peptide fragments (represented by dashed lines) were not analyzed for their N-terminal sequences. The asterisk represents the residue at position 2 of the CBA subunit, where a different amino acid was detected between the sequence deduced from the cDNA and the determined sequence by Edman degrada-tion. The signal peptide region is underlined.
Absorbance at 280 nm
Absorbance at 220 nm
B
A
0 10 20 30 40 50 60
0 10 20 30 40 50 60
Acetonitrile (%)
Retention time (min)
L5
L4 L3
L2 L1 0.05
0
0.2
0 0.4
Retention time (min)
Acetonitrile (%)
Fig. 2. Reverse-Phase HPLC of rPE-CBA and Its Peptide Fragments Generated by Enzymatic Digestion.
four
C-terminal
amino
acids
(-Asn-Met-Asp-Thr,
462 Da) of the subunit polypeptide were
post-transla-tionally truncated to afford the mature subunit. The
mature subunit contained seven cysteine residues
(Fig. 3B). It was concluded from these data that CBA
consisted of an SS-linked homodimer of a 9257-Da
polypeptide (84 amino acids). No significantly similar
sequences to the CBA subunit could be found during an
in silico
search, indicating CBA to be a novel protein.
Discussion
We isolated in this study a novel lectin from the green
alga,
C. barbatum
, which we call
Codium barbatum
lectin (CBA). The hemagglutination-inhibition profile of
CBA was obviously distinct from those of the
N
-acetylgalactosamine-specific lectins previously isolated
from other species belonging to this genus,
22–24)as the
activity of CBA was not inhibited by
N
-acetylgalactos-amine or
N
-acetylglucosamine. CBA is therefore the
first example among the
Codium
lectins having no
affinity for monosaccharides. However, the activity was
inhibited by PTG and remarkably further inhibited by
asialo-PTG (Table 3), suggesting that the branched
sugar moiety of PTG was responsible for this inhibition.
With respect to the glycan structure, PTG is known to
have two types of
N
-glycans in the molecule: a complex
type and high-mannose type.
33–35)Yeast mannan, which
contains only high-mannose type
N
-glycans, was not
inhibitory, leading to the supposition that CBA
recog-nized the complex type of
N
-glycans. However,
trans-ferrin and fetuin bearing the complex type of
N
-glycans
were also not inhibitory. This discrepancy may be
derived from a difference in the number of core (1,6)
fucose residues of the complex type of
N
-glycans
between PTG and transferrin or fetuin. We therefore
speculate that CBA could recognize core (
1,6)
fucosy-lated
N
-glycans that are predominant in PTG, but are
few or absent in transferrin and fetuin.
36)Details of the
carbohydrate-recognition mode, including the
oligosac-charide-binding specificity, of CBA are now under
investigation in our laboratory.
As the first example among
Codium
lectins, we have
elucidated the primary structure of CBA by a
combina-tion of ESI-MS, Edman degradacombina-tion, and cDNA cloning.
CBA consisted of an SS-linked homodimer of a
9257-Da subunit and contained a total of 14 half-cystines
involved in disulfide linkages. A potential
N
-glycosyla-tion site (Asn
83-Gly
84-Ser
85) was found in the sequence
of the CBA subunit, although the presence of the
consensus peptides (Asn-X-Thr/Ser) does not always
lead to glycosylation.
37)In addition, the determined
molecular mass of the peptide fragment (L5) containing
the triplet sequence of Asn
83-Gly
84-Ser
85coincided with
the calculated mass from the sequence, suggesting that
CBA had no carbohydrate. Serine was deduced at the
second residue of the CBA subunit from cloning,
whereas threonine was determined by Edman
degrada-tion. The presence of a different amino acid at position 2
of the CBA subunit suggests that isoforms of the CBA
subunits exist, or polymorphism between the local
populations of Satsuma-Iwojima Island and Tanegashima
Island, where the algal samples were respectively
collected for lectin purification and RNA extraction
(see the Materials and Methods section). The occurrence
of isolectins has also been reported for the red alga,
Hypnea japonica
.
15,16)No significantly similar proteins were found,
includ-ing lectins, durinclud-ing
in silico
searches for the amino acid
sequences. The amino acid sequence of CBA is
there-fore distinct from that of land plant lectins, and also
from the monosaccharide-binding lectins of other green
algae like
Enteromorpha prolifera
19)and
Ulva pertusa
21)that are respectively specific for
L-fucose/
D-mannose
and
N
-acetylglucosamine. Although there is no
infor-mation on the amino acid sequences of
Codium
lectins,
several lectins from
Codium
, including those isolated
from
C. fragile
ssp.
tomentosoides
,
22)C. fragile
ssp.
atlanticum
,
22)C. tomentosum
,
23)and
C. giraffa
,
24)have
been partially characterized. CBA clearly differed from
known
Codium
lectins with respect to such biochemical
properties as the molecular mass, amino acid
composi-tion, and carbohydrate-binding specificity, indicating
CBA to be a novel lectin. It would be of interest to
elucidate and compare the primary structures of lectins
from other species of
Codium
, because about 18 species
of the genus have been reported in Japan.
38)The present study has demonstrated the interesting
proposition that the four amino acids in the C-terminal
region of the CBA precursor could be
post-translation-ally truncated. Many lectins contain propeptide regions
at the N- and/or C-termini of their precursors, and the
propeptides are truncated in the mature form.
1–4)Some of
the propeptide regions have been reported to be required
for the subcellular localization and inactivation of lectins
in a temporarily inappropriate location.
39)For instance,
Galanthus nivalis
(snowdrop) lectin (GNA) is
synthe-sized on the endoplasmic reticulum as a preproprotein
which contains a signal peptide and a C-terminal
propeptide,
40)and both the pre- and pro-peptides of the
lectin are necessary for trafficking to the vacuole.
39)Wheat germ agglutinin (WGA) is also processed and
sorted to the vacuole in a similar fashion,
41,42)although
the two lectins, GNA and WGA, are structurally and
functionally distinct from each other. A lectin (BCA),
which recognized the cluster of non-reducing terminal
(1,2) mannose residues of high-mannose
N
-glycans, has
recently been isolated from the green alga,
Boodlea
coacta
, and its precursor also contained a propeptide at
the C-terminus.
13)The recombinant of the mature form of
BCA presented hemagglutination activity, whereas the
pro-form did not show this activity (details of these
results will appear elsewhere), suggesting that the
propeptide regions of algal lectin precursors could also
control their hemagglutination activity and, possibly,
their subcellular localization. The sequence of the
propeptide from CBA was quite different from the
known subcellular localization signals of both lectins and
of other proteins.
43)Further investigations are required to
elucidate such functions of the propeptide as preparing
the recombinants of mature- and prepro-forms of CBA,
and to examine the hemagglutination activities and
subcellular localizations of these forms in the alga.
Acknowledgments
and to Dr. Koji Muramoto (Tohoku University) for
providing the silica gel-immobilized phosphorylcholine
(PC 300S (N)) column. We thank Dr. Tomoko Amimoto
in the Natural Science Center for Basic Research and
Development (N-BARD) at Hiroshima University for
measuring the ESI-MS data. We also thank Dr.
Lawrence M. Liao (Hiroshima University) for a critical
reading of the manuscript. This work was partially
supported by the Program for Promotion of Basic and
Applied Researches for Innovations in Bio-Oriented
Industry of Japan.
References
1) Sharon N and Lis H, ‘‘Lectins’’ 2nd ed., Kluwer Academic Publisher, Dordrecht, pp. 1–454 (2003).
2) van Damme EJM, Peumans WJ, Barre A, and Rouge P,Crit.
Rev. Plant Sci.,17, 575–692 (1998).
3) Lis H and Sharon N,Chem. Rev.,98, 637–674 (1998). 4) Dodd RG and Drickmer K,Glycobiology,11, 71–79 (2001). 5) Ohno Y, Naganuma T, Ogawa T, and Muramoto K,J. Agric.
Food Chem.,54, 548–553 (2006).
6) Boyd MR, Gustafson KR, Mcmahon JB, Shoemaker RH, O’Keefe BR, Mori T, Gulakowski RJ, Wu L, Rivera MI, Laurencot CM, Currens MJ, Cardellina JH II, Buckheit RWJr, Nara PL, Pannell LK, Sowder RC II, and Henderson LE,
Antimicrob. Agents Chemother.,41, 1521–1530 (1997).
7) Bokesch HR, O’Keefe BR, McKee TC, Pannell LK, Patterson ML, Gardella RS, Sowder RC II, Turpin J, Watson K, and Buckheit RWJr,Biochemistry,42, 2578–2584 (2003). 8) Bewley CA, Cai M, Ray S, Ghirlando R, Yamaguchi M, and
Muramoto K,J. Mol. Biol.,339, 901–914 (2004).
9) Mori T, O’Keefe BR, Sowder RC II, Bringans S, Gardella RS, Berg S, Cochran P, Turpin JA, Buckheit RWJr, McMahon JB, and Boyd MR,J. Biol. Chem.,280, 9345–9353 (2005). 10) Hori K, Sato Y, Ito K, Fujiwara Y, Iwamoto Y, Makino H, and
Kawakubo A,Glycobiology,17, 479–491 (2007).
11) Sato Y, Okuyama S, and Hori K,J. Biol. Chem.,282, 11021– 11029 (2007).
12) Sato Y, Hirayama M, Morimoto K, and Hori K, Biochem.
Biophys. Res. Commun.,405, 291–296 (2011).
13) Sato Y, Hirayama M, Morimoto K, Yamamoto N, Okuyama S, and Hori K,J. Biol. Chem.,286, 19446–19458 (2011). 14) Hung LD, Sato Y, and Hori K,Phytochemistry,72, 855–861
(2011).
15) Hori K, Matsubara K, and Miyazawa K, Biochim. Biophys.
Acta,1474, 226–236 (2000).
16) Okuyama S, Nakamura-Tsuruta S, Tateno H, Hirabayashi J, Matsubara K, and Hori K,Biosci. Biotechnol. Biochem., 73, 912–920 (2009).
17) Fukuda Y, Sugawara T, Ueno M, Fukuta Y, Ochi Y, Akiyama K, Miyazaki T, Matsuda S, Kawakubo A, and Kato K,
Anticancer Drugs,17, 943–947 (2006).
18) Ziolkowska NE and Wlodawer A,Acta Biochim. Pol.,53, 617– 626 (2006).
19) Ambrosio AL, Sanz L, Sanchez EI, Todel CW, and Calvete JJ,
Arch. Biochem. Biophys.,415, 245–250 (2003).
20) Sampaio AH, Rogers DJ, and Barwell CJ,Bot. Mar.,41, 427– 433 (1998).
21) Wang S, Zhong FD, Zhang YJ, Wu ZJ, Lin QY, and Xie LH,
Acta Biochim. Biophys. Sin.,36, 111–117 (2004).
22) Rogers DJ, Loveless EW, and Balding P, ‘‘Lectins: Biology, Biochemistry, Clinical Biochem.,’’ Walter de Gruyter, Berlin, pp. 155–160 (1986).
23) Fabregas J, Mun˜oz A, Llovo J, and Carracedo A,J. Exp. Mar.
Biol. Ecol.,124, 21–30 (1988).
24) Alvarez-Hernandez S, de Lara-Isassi G, Arreguin-Espinoza R, Arreguin B, Hernandez-Santoyo A, and Rodriguez-Romero A,
Bot. Mar.,42, 573–580 (1999).
25) Wu AM, Song SC, Chang SC, Wu JH, Chang KS, and Kabat
EA,Glycobiology,7, 1061–1066 (1997).
26) ‘‘Seaweed Resources of the World,’’ eds. Critchley AT and Ohno M, Japan International Cooperation Agency, Yokosuka, pp. 1–431 (1998).
27) Hori K, Miyazawa K, Fusetani N, Hashimoto K, and Ito K,
Biochim. Biophys. Acta,873, 228–236 (1986).
28) Watanabe Y, Abolhassani M, Tojo Y, Suda Y, Miyazawa K, Igarashi Y, Sakuma K, Ogawa T, and Muramoto K,
J. Chromatogr. A,1216, 8563–8566 (2009).
29) Hori K, Miyazawa K, and Ito K,Bull. Jpn. Soc. Sci. Fish.,52, 323–331 (1986).
30) Lowry OH, Rosebrough NJ, Farr AL, and Randall RJ,J. Biol.
Chem.,193, 265–275 (1951).
31) Scha¨gger H and von Jogow G,Anal. Biochem.,166, 368–379 (1987).
32) Bendtsen JD, Nielsen H, von Heijne G, and Brunak S,J. Mol.
Biol.,340, 783–795 (2004).
33) Tsuji T, Yamamoto K, Irimura T, and Osawa T,Biochem. J.,
195, 691–699 (1981).
34) Yamamoto K, Tsuji T, Irimura T, and Osawa T,Biochem. J.,
195, 701–713 (1981).
35) de Waard P, Koorevaar A, Kamerling J, and Vliegenthart JFG,
J. Biol. Chem.,266, 4237–4243 (1991).
36) Spik G, Bayard B, Fournet B, Strecker G, Bouquelet S, and Montreuil J,FEBS Lett.,50, 296–299 (1975).
37) Gavel Y and von Heijne G,Protein Eng.,3, 433–442 (1990). 38) Verbruggen H, Leliaert F, Maggs CA, Shimada S, Schils T,
Provan J, Booth D, Murphy S, Clerck O, Littler DS, Littler MM, and Coppejans E,Mol. Phylogenet. Evol.,44, 240–254 (2007).
39) Fouquaert E, Hanton SL, Brandizzi F, Peumans WJ, and van Damme EJM, Plant Cell Physiol., 48, 1010–1021 (2007).
40) van Damme EJM, Kaku H, Perini F, Goldstein IJ, Peeters B, Yagi F, Decock B, and Peumans WJ,Eur. J. Biochem.,202, 23– 30 (1991).
41) Mansfield MA, Peumans WJ, and Raikhel NV,Planta, 173, 482–489 (1988).
42) Bednarek SY, Wilkins TA, Dombrowski JE, and Raikhel NV,
Plant Cell,2, 1145–1155 (1990).