Endoreduplication, a strategy to amplify nuclear DNA without cell division, is very common but poorly understood in plants. Recent findings in Drosophilaprovide a first picture of the molecular mechanism, which appears to be conserved between plants and animals. In Arabidopsis, the study of trichomes, leaf epidermis and hypocotyl cells sheds new light on the developmental regulation of this process, and its relation to cell expansion.
Addresses
Laboratoire de Biologie Cellulaire, INRA, Route de Saint-Cyr, 78026 Versailles cedex, France
*e-mail: [email protected] ‡e-mail: [email protected] §e-mail: [email protected]
†Universität Tübingen, Auf der Morgenstelle 1, D-72076 Tübingen, Germany; e-mail: [email protected]
Current Opinion in Plant Biology1998, 1:498–503 http://biomednet.com/elecref/1369526600100498 © Current Biology Ltd ISSN 1369-5266
Abbreviation
CDK cyclin dependent kinase
Introduction
Endoreduplication involves one or several rounds of nuclear DNA synthesis without chromosomal and cellular division and leads to polyploidy. It is widespread among eukaryotes and estimated to occur in over 90% of all angiosperms [1,2]. A variety of biological processes, includ-ing cell differentiation, cell expansion, metabolic activity, and resistance against irradiation have been proposed to involve endoreduplication [1–3]. The evidence for these claims, however, remains circumstantial and both the role and control of the endocycle are poorly characterised in plants. Fortunately, recent progress has opened new per-spectives. In this review, we will discuss these advances on the understanding of the function and regulation of endoreduplication in higher plants, with emphasis on developmental aspects. Where relevant we will refer to work on Drosophila, which is at present the model system where the endocycle has been best characterised.
Mitosis and endoreduplication: two different
cycles with a common molecular basis
Endoreduplication shares several characteristics with the mitotic cycle which is controlled by a set of highly con-served kinases and phosphatases. At the core of this ‘machinery’ are the cyclin dependent kinases (CDKs), which need to bind to the regulatory cyclins in order to become activated. Functionally distinct cyclins and CDKs exist which in different combinations control the transi-tions from one cell cycle phase to the other. For instance,
the so called D-type and E-type cyclins are thought to be necessary for the transition from G1 to S, whereas the B-cyclins function during the G2 to M transition. The endoreduplicative cycle, which has been less well charac-terised, appears to be under control of the same type of complexes, as was shown for Drosophila[4]. Although they have a common molecular basis, it is obvious that both cycles are mutually exclusive and higher eukaryotes have developed strategies which assure an inhibition of endoreduplication during mitosis and vice versa.
Components that repress endoreduplication have been identified in different eukaryotes. In Drosophila, ESCAR-GOT, a potential transcription factor, appears to be required to keep imaginal disk cells in a mitotic state [5]. In addition, the myb gene DmMyb is likely to function in both the stimulation of mitosis and the repression of endoreduplication [6].
How do cells switch from mitosis to endoreduplication? Although this process is still poorly understood, it is clear that it requires important changes in the cell cycle machin-ery. For instance, cyclins that are active in G2 are no longer expressed, and others have modified activities as was reported for E cyclins in Drosophila: E cyclin is present at constant levels in mitotic cells, but fluctuating levels are required for multiple rounds of endocycles [4]. One of the components necessary for the transition to endoreduplica-tion is the FIZZY RELATED protein, first identified in Drosophila. It was proposed that FIZZY RELATED func-tions in the degradation and inactivation of mitotic cyclins during interphase, thus allowing endoreduplication to occur [7].
Recent investigations suggest that the regulation of endoreduplication in plants is similar to that in animals. In particular, biochemical analysis of endoreduplication in maize has revealed the existence of an as of yet uncharac-terised factor, which inhibits the kinase activity of the mitotic CDK/cyclin complex [8]. This implies that in plants as well as Drosophila, the transition to endoredupli-cation requires downregulation of the mitotic cycle. In the maize system, a member of the retinoblastoma family, which in animals controls cell proliferation and possibly also endoreduplication, has been identified [9,10]. The exact role of this protein remains to be established. E-cyclins have not been found, although a member of the D-cyclin family in plants, has some characteristics in com-mon with the E-cyclins [11]. A role for the Arabidopsis CDK cdc2bAT in endoreduplication has been proposed, as its mRNA was detected in endoreduplicating cells [12]. Homologs of the FIZZY RELATED gene are present in the Arabidopsis genome (J Traas, H Höfte, unpublished data), although their function has not yet been studied.
Endoreduplication and development: rule without dividing?
Switching from mitosis to endoreduplication:
developmental and environmental regulation
The molecular data summarised above indicate that the basic mitotic machinery is the immediate target for the switch from mitosis to endoreduplication. How is this switch regulated; what are the signals that induce such changes during development? The transition to endocy-cles is subject to regulation by developmental cues, and other endogenous and environmental signals [1,2,13]. The relative contribution of these cues, however, varies consid-erably in different cell types. To illustrate this, we will briefly describe the developmental control of the switch to endoreduplication for three different cell types in Arabidopsis: hypocotyl cells [14•], leaf epidermal pavement cells [15], and trichomes [16].
Most of the initial growth of the seedling is due to an enor-mous elongation of the hypocotyl. Before germination, all cells of the embryo have a ploidy level of 2C. One day after the induction of germination, before any significant cell expansion occurs, between 12 and 43% of the cells have
undergone one or two endoreduplication cycles. Finally, up to 80% of all hypocotyl cells become polyploid. Strikingly, virtually no cell divisions take place in the endodermis or in the cortex and mitotic division in the epi-dermis is restricted to the forming stomata. Thus, at the end of embryogenesis, the hypocotyl apparently contains differentiated, non-mitotic cells that are programmed to enter endoreduplication immediately upon induction of seed germination (Figure 1a).
Leaf epidermal tissues in Arabidopsiscontain three differ-ent cell types: guard cells, trichomes and pavemdiffer-ent cells. The differentiated pavement cells, easily recognised by their puzzle-like form, show a DNA content that varies between 2C and 16C. Approximately 21% of these cells, however, still show a DNA content of 2C [15]. This rules out the idea that pavement cells require endoreduplication for cell differentiation. Endoreduplication is rather one optional aspect of pavement cell differentiation which probably is regulated by external cues.
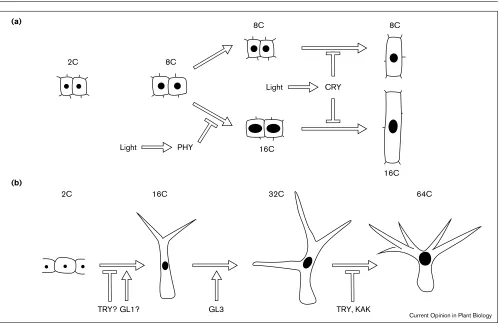
Figure 1
2C
2C
8C
8C
16C
CRY Light
16C 32C 64C
16C 8C
Light PHY
TRY? GL1? GL3 TRY, KAK
(a)
(b)
Current Opinion in Plant Biology
Factors involved in the control of endoreduplication in differentiating cells of Arabidopsis.(a)The control of endoreduplication in hypocotyl cortical and epidermal cells. These cells initially have a DNA level of 2C. Upon germination, two rounds of endoreduplication lead to the formation of a population of 8C cells. In the dark, a third endocycle takes place. This cycle is inhibited by light via phytochrome (PHY). Cryptochrome (CRY) does not interfere with endoreduplication, but is necessary for the inhibition of growth by blue light. (b)The control of
This is different for trichomes, which are large single, branched cells that originate from protodermal cells. On average mature trichomes show an average DNA content of 32C suggesting that they undergo four cycles of endoreduplication (Figure 1b). By morphological criteria, incipient trichomes can first be recognised by an increase in nuclear DNA content, due to the first two endocycles. Thus, the mitosis–endoreduplication switch appears to be an integral part of the trichome differentiation program. Does this also mean that this switch has a role in the cell fate decision? Genetic evidence supports this possibility since two genes, GLABRA1 (GL1) and TRIPTYCHON (TRY), known to have a role in trichome patterning, are also involved in the regulation of endoreduplication [16,17]. GL1 is a positive regulator of trichome develop-ment, and plants overexpressing the gene exhibit an additional round of endoreduplication in mature tri-chomes. TRY is a negative regulator of trichome development, and trymutant trichomes contain twice the DNA content of wild-type. Thus, a positive or negative effect on trichome development is correlated with respec-tively a positive or negative effect on endoreduplication. Although other explanations are possible, an exciting sce-nario is that trichome fate determination is directly triggered via the cell cycle machinery. Such a direct link between cell fate and the cell cycle has been proposed for germ-line cysts in Drosophila[18]. New cysts are formed by a series of incomplete cell divisions, resulting in sixteen interconnected cells. One of these cells will differentiate as an oocyte, whereas the others will become nurse cells and immediately initiate a series of endocycles. These endocy-cles are tightly coupled to cell differentiation. Interestingly, a viable cyclin E mutant shows a defect in endoreduplication and a defect in cell differentiation; the number of cells that will become oocytes is increased, indi-cating that cell differentiation is directly modified through a change in cell cycle regulation.
In the three examples, hypocotyl cells, pavement cells, and trichomes, the developmental restrictions for the switch from mitotic division to endoreduplication are very different. Whereas during trichome development the switch from mitotic divisions to endoreduplication appears to be an invariant part or a direct consequence of the cell fate decision, certain hypocotyl cells are pro-grammed to enter endoreduplication but remain in a quiescent state until they receive an additional signal dur-ing germination. Epidermal pavement cells may or may not enter endoreduplication cycles, the actual switch in this cell type is apparently governed by endogenous or environmental cues.
The number of endocycles: developmental and
environmental control
Differentiating cells can undergo multiple cycles of endoreduplication, which in some cases can result in a massive amplification of the amount of DNA per cell. This is spectacularly illustrated by endosperm cells of
Arum maculatum which contain nuclei of up to 24,576C
[19]. The number of endoreduplication cycles appears to be under the control of both developmental and environ-mental signals.
In certain cell types, the number of endocycles is strictly correlated with the differentiation program. This is well-studied in Arabidopsistrichomes where several positive or negative regulators of the number of endocycles have been identified. The GLABRA3(GL3) gene is required to pro-ceed from the third to the fourth endocycle, whereas KAKTUS (KAK) [20•], functions to limit the number of cycles. Similarly to trymutants, kaktrichomes contain, on average, twice the DNA of wild-type hair cells. Although the specificity of different genes for distinct endoredupli-cation steps suggests a stepwise regulation of endoreduplication, it is equally possible that the products of these genes act in a concentration-dependent manner.
Ploidy levels can also be regulated by environmental sig-nals. A factor often associated with the control of ploidy levels is light. This was confirmed recently for the Arabidopsis hypocotyl, where the third endocycle is inhib-ited by light, through the action of the red light/far red light photoreceptor phytochrome [14•]. It was shown that 16C nuclei are absent in wild type hypocotyls grown under continuous far red light, and present in hypocotyls of phy-tochrome (phy) Amutants.
Cell volume and ploidy levels
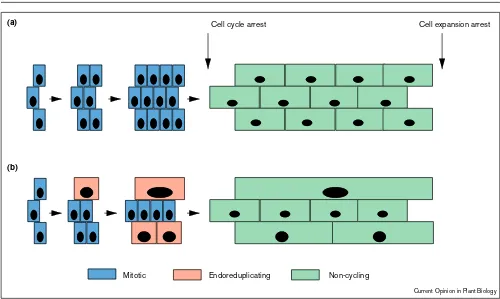
Figure 2
Mitotic Endoreduplicating Non-cycling
Cell cycle arrest Cell expansion arrest
Current Opinion in Plant Biology (a)
(b)
Scenario explaining the correlation between ploidy levels and cell size within a given tissue. An expanding organ can be considered as being composed of fields of cells that simultaneously react to developmental cues. During the life time of a cell, two consecutive developmental signals provoke such a simultaneous response, a first one triggering the arrest of the cell cycle, and a second one triggering the arrest of cell expansion. These signals may be endogenous positional cues causing the distinct zonation that can be observed in roots or leaves [26•,27•], or environmental cues, such as light during hypocotyl development [14•]. (a)In some tissues, cells never endoreduplicate. Upon receiving the cell cycle arrest signal, these cells will increase in size until cell
expansion is arrested simultaneously. This results in a homogeneous final cell size distribution within the tissue, as observed, for instance, in sunflower leaves [27•]. (b)In other tissues, dividing cells can switch to endoreduplication. As discussed in the text, this switch is not necessarily synchronous for neighbouring cells. As a result, more or less important size differences will have built up by the time the cells receive the cell cycle arrest signal. These differences will be further amplified during the cell expansion phase, and a result, the final cell size will be correlated with the ploidy levels. Note also that at the end of the growth phase the ratio between the amount of DNA and the total tissue mass is conserved whether or not endoreduplication takes place.
Figure 3
Non-synchronous switch from cell division to endoreduplication in a leaf primordium. (a)
Low magnification, showing both nuclear (red) and cytoplasmic (green) staining and (b)
higher magnification showing nuclear staining using propidium iodide only. A trichome precursor cell in an Arabidopsisleaf primordium has stopped dividing and switched to endoreduplication (note the larger nucleus (arrowhead)) while surrounding cells are still actively dividing (arrows). Scale bars represent 20µm (a) and 10µm (b).
Current Opinion in Plant Biology
the nucleus. How the correlation between cell size and DNA-levels is established in such a context is not known, but a possible scenario is presented in Figures 2 and 3.
If DNA levels do not strictly determine the dimensions of individual cells, how should we interpret the function of the intimate link with size? In general terms, one role of endoreduplication, like for mitosis [24], can be understood as a mechanism to generate sufficient DNA to support a future massive increase in tissue mass. Rather than prede-termine final cell size, specific endoreduplication levels would allow a more or less loosely defined range of cell vol-umes to be defined. The exact size of a group of cells would be determined afterwards by other factors. This is clearly illustrated by the observation that the same cell with the same DNA content can reach different sizes depending on the environment. Indeed, hypocotyls bear-ing a mutation in the blue light/UV-A receptor CRYPTOCHROME1, grown in blue light are three times longer than those of wild-type seedlings, but do not show altered ploidy levels, indicating that this receptor mediates the inhibition of cell elongation, without affecting the number of endocycles [14•]. Endoreduplication might also promote cell size diversity by allowing a selective cell divi-sion arrest. This strategy may have a selective advantage since only in this way can the same ratio between nuclear and cellular size be maintained in all cells within an organ, which in theory should favour the metabolic homeostasis within the tissue (for a discussion of this aspect in mam-malian liver cells see [25]). The biological function of producing a large cell instead of several smaller cells, may be more (as in xylem cells) or less (as in epidermal cells) obvious and the answer to this question will vary depend-ing on the cell- and tissue type investigated.
Conclusions
In conclusion, recent developments in Drosophila have provided new insights into the molecular mechanism of endoreduplication and have identified genes involved in the regulation of this process. Homologues for some of these genes are also present in plants, creating the oppor-tunity to investigate their role in endoreduplication. This may be achieved using reverse genetics in Arabidopsis, and eventually may allow this process to be manipulated in crop plants. Several genes regulating trichome differentia-tion and endoreduplicadifferentia-tion have been identified by mutation in Arabidopsis. This shows that the process is amenable to genetic dissection, which should open up important new perspectives.
The study of developmental processes in which endo-ploidy occurs shows that endoreduplication takes place early during development, and that it is tightly coupled to cell differentiation. In all cases investigated it is also inti-mately linked to cell expansion. The existing evidence suggests that endoreduplication is not a regulator for cell expansion, but rather allows for the amplification of nuclear DNA in anticipation of a massive expansion.
Acknowledgements
We would like to thank Valérie Gaudin, Heather McKhann, Fabien Nogué and Téva Vernoux for helpful discussions and correction of the manuscript. Olivier Grandjean is thanked for help in making Figure 3.
References and recommended reading
Papers of particular interest, published within the annual period of review, have been highlighted as:
• of special interest
••of outstanding interest
1. Barlow P:Endopolyploidy: towards an understanding of its biological significance.Acta Biotheor 1978, 27:1-18. 2. D’Amato F:Polyploidy in cell differentiation. Caryologia 1989,
42:183-211.
3. Nagl W: DNA endoreduplication and polyteny understood as evolutionary strategies. Nature1976, 261:614-615. 4. Weiss A, Herzig A, Jacobs H, Lehner C:Continuous Cyclin E
expression inhibits progression through endoreduplication cycles in Drosophila. Curr Biol 1998, 8:239-242.
5. Fuse N, Hirose S, Hayashi S:Diploidy of Drosophiliaimaginal cells is maintained by transcriptional repressor encoded by escargot.
Genes Dev 1994, 8:2270-2281.
6. Katzen AL, Jackson J, Harmon BP, Fung SM, Ramsay G, Bishop JM:
Drosophila myb is required for the G2/M transition and maintenance of diploidy.Genes Dev1998, 12:831-843. 7. Sigrist SJ, Lehner CF: Drosophila fizzy-relateddown-regulates
mitotic cyclins and is required for cell proliferation arrest and entry into endocycles. Cell1997, 90:671-681.
8. Grafi G, Larkins B:Endoreplication in Maize endosperm:
involvement of M phase-promoting factor inhibition and induction of S phase-related kinase. Science 1995, 269:1262-1264. 9. Grafi G, Burnett RJ, Helentjaris T, Larkins BA, DeCaprio JA, Sellers
WR, Kaelin WG Jr: A maize cDNA encoding a member of the retinoblastoma protein family: involvement in endoreduplication.
Proc Natl Acad Sci USA1996, 93:8962-8967.
10. Niculescu AB III, Chen X, Smeets M, Hengst L, Proves C, Reed SI:
Effects of p21(Cip1/Waf1) at both the G1/S and G2/M cell cycle transitions: pRb is a critical determinant in blocking DNA replication and in preventing endoreduplication.Mol Cell Biol
1998, 18:629-643.
11. Soni R, Carmichael J, Shah Z, Murray J:A family of cyclin D homologs from plants differentially controlled by growth regulators and containing the retinoblastoma protein interaction motif.Plant Cell 1995, 7:85-103.
12. Segers G, Gadisseur I, Bergounioux C, de Almeida Engler J, Jacqmard A, van Montagu M, Inzé D:The Arabidopsis cyclin-dependent kinase gene cdc2bAT is preferentially expressed during S and G2 phases of the cell cycle.Plant J1996, 10 :601-612.
13. Smith A, Orr-Weaver T: The regulation of the cell cycle during Drosophilaembryogenesis : the transition to polyteny.
Development1991, 112:997-1008.
14. Gendreau E, Höfte H, Grandjean O, Brown S, Traas J:Phytochrome
• controls the number of endoreduplication cycles in the Arabidopsis thaliana hypocotyl.Plant J 1998, 13:221-230. In this paper it is shown that phytochrome specifically inhibits a third endoreduplication cycle in developing hypocotyl cells, illustrating that envi-ronmental factors can influence ploidy levels. It is also shown that cryp-tochrome controls cell elongation, without affecting endoreduplication. 15. Melaragno JE, Mehrotra B, Coleman AW:Relationship between
endopolyploidy and cell size in epidermal tissue of Arabidopsis.
Plant Cell 1993, 5:1661-1668.
16. Hülskamp M, Misera S, Jürgens G:Genetic dissection of trichome cell development in Arabidopsis. Cell 1994, 76:555-566. 17. Larkin J, Oppenheimer D, Lloyd A, Paparozzi E, Marks M:Roles of
GLABROUS1and TRANSPARENT TESTA GLABRAin Arabidopsis trichome development. Plant Cell 1994, 6:1065-1076.
18. Lilly A, Spradling A: The Drosophilaendocycle is controlled by cyclin E and lacks a checkpoint ensuring S-phase completion.
19. Erbrich P:Uber Endopolyploidie und kernstrukturen in Endospermhaustorien. Oester Bot Z 1965, 112:197-262. [Title translation: about endopolyploidy and nuclear structures in endosperm haustorien.]
20. Folkers U, Berger J, Hülskamp M:Cell morphogenesis of trichomes
• in Arabidopsis: differential control of primary and secondary branching by branch initiation regulators and cell growth.
Development1997, 124:3779-3786
This paper describes a genetic analysis of cell expansion and branching dur-ing trichome formation. Mutations that cause an increase (kak, try) or decrease (gl3) in cell size and branching also cause a increase or decrease in ploidy levels.
21. Cavallini A, Natali L, Cionini G, Balconi C, D’Amato F:Inheritance of nuclear DNA content in leaf epidermal cells of Zea maysL.Theor Appl Genet 1997, 94:782-787.
22. Cionini PG, Cavallini A, Baroncelli S, Lercari B, D’Amato F: Diploidy and chromosome endoreduplication in the development of epidermal cell lines in the first foliage leaf durum wheat (Triticum durumDesf.).Protoplasma1983, 118:156-162.
23. List A:Some observations on DNA content and cell and nuclear volume growth in the developing xylem cells of certain higher plants. Am J Bot 1963, 50:320-329.
24. Smith L: What is the role of cell division in leaf development?Sem Cell Dev Biol 1996, 7:839-848.
25. Epstein CJ: Cell size, nuclear content, and the development of polyploidy in the mammalian liver.Proc Natl Acad Sci USA 1967,
57:327-334.
26. Jacobs T:Why do plant cells divide?Plant Cell 1997,
• 9:1021-1029.
A well written review on cell cycle control in plants. The relationship between cell expansion and cell division is extensively discussed.
27. Granier C, Tardieu F: Spatial and temporal analyses of expansion