Accumulating evidence suggests that lateral transfer of nodulation capacity is an important driving force in symbiotic evolution. As a consequence, many distantly related soil bacteria have acquired the capacity to invade plants and fix nitrogen within them. In addition to these proteins required for bacteroid development and nitrogen fixation, core symbiotic competence seems to require flavonoids, NodD proteins, lipochitooligosaccharidic Nod-factors, extra-cellular polysaccharides, as well as various exported proteins. Plants respond to different levels and combinations of these substances in species specific ways. After contact has been initiated by flavonoids and NodD proteins, constant signal exchange fine-tunes these symbiotic demands, especially to overcome defence reactions.
Addresses
Laboratoire de Biologie Moléculaire des Plantes Supérieures (LBMPS), Université de Genève, 1 ch. de l’Impératrice, 1292 Chambésy/Genève, Switzerland
*e-mail: [email protected]
Current Opinion in Plant Biology1999, 2:305–311 http://biomednet.com/elecref/1369526600200305 © Elsevier Science Ltd ISSN 1369-5266
Abbreviations
Hac root-hair curling
Had root-hair deformation
Hai root-hair formation
Introduction
In the latter part of the nineteenth century, inquiry into the nature of plant–microbe interactions turned from the observational to the experimental. Beyerinck proved that bacteria elicit nodules on the roots of legumes by preparing pure cultures from nodules of Vicia faba and using them to infect Faba beans growing in sterile soil [1]. Prazmowski [2] inoculated Pisum sativumwith axenic cultures and observed that micro-organisms entered into the plant via infection threads in root-hairs. Hiltner [3] prepared aqueous, bacteria-free filtrates from mature P. sativumnodules and demonstrated that they contain a substance that induces root-hair formation (Hai) and deformation of the root-hairs (Had). Ninety years later Lerouge et al. [4] showed that the root-hair deforming substances of R. meliloti are N-acylated oligomers of N-acetyl-D-glucosamine (known as Nod-factors) and implied that variations in Nod-factor structure are der-minants of host specificity. In turn, Spaink et al. [5] demonstrated that the principal differences between the Nod-factors of R. leguminosarumbiovar viciaeand those of R. meliloti lie in the degree of unsaturation of the acyl chains, as well as the presence or absence of a sulphate group on the reducing-terminal glucosamine.
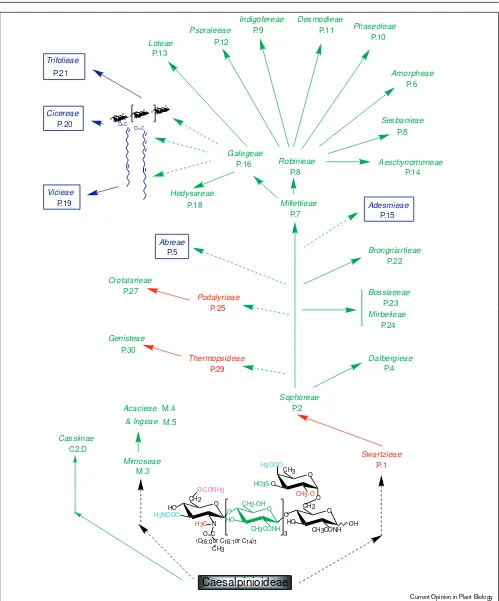
Common threads link these milestones in symbiotic nitro-gen fixation research — all were performed by European scientists on local legumes and their associated rhizobia. Unfortunately, the plants they used (Medicago, Pisum, and Vicia) are very closely related and belong to only two (Vicieaeand Trifolieae) of the thirty-nine recognised tribes (three of the 666 genera) of the Leguminosae (see [6] and Figure 1). Furthermore, their respective symbionts, R. meliloti, R. leguminosarumbv. viciaeand R. leguminosarum bv. trifolii, associate with only few plants (i.e. have a narrow host-range). It is thus possible that nitrogen-fixing sym-bioses in temperate, herbaceous legumes are not representative of Rhizobium–legume associations as a whole. To seek a more general understanding of the mole-cular bases of symbioses, we have examined the relationship between promiscuous (i.e. able to nodulate many different legumes) rhizobia and their hosts. This raises the question of rhizobial taxonomy. Below, we will argue that rhizobia are heterogenous because, like other Gram-negative bacteria, they have acquired large blocks of genetic information by lateral-transfer. In some cases these modules represent pathogenicity islands, whereas in others they carry symbiotic genes (see note added in proof). Once acquired these ‘symbiotic islands’ have been subject to constant re-arrangements, amplifications and deletions.
Rhizobia: phylogenetically distant bacteria
containing related symbiotic genes
Hiltner and Störmer [7] were probably the first to attempt classification of rhizobia on the basis of their plant origins. Host-based classifications would work if rhizobial host-ranges mirrored legume taxonomy, but some rhizobia form symbioses across taxonomic divisions. Wilson [8], for exam-ple, tested the host-ranges of rhizobia isolated from 31 different genera of legumes on 160 different legume species. The average number of plant species nodulated by a particular isolate was 33%, and four of the isolates formed nodules on more than half the legumes tested. Likewise, Rhizobium sp. NGR234 nodulates at least 112 genera of legumes, whereas the closely related R. fredii USDA257 nodulates an exact subset (77 genera) of the NGR234 hosts [9•]. Widespread existence of such broad host-range
rhizo-bia, as well as promiscuous hosts such as Vigna[10] renders any host-based classification scheme inadequate.
Figure 1
Caesalpinioideae
MimoseaeCassiinae
Acacieae & Ingeae
Swartzieae Dalbergieae Thermopsideae
Genisteae
Mirbelieae
Podalyrieae Bossiaeeae
Crotalarieae
Abreae
Brongniartieae Millettieae Adesmieae
Robinieae Aeschynomeneae
Galegeae
Hedysareae
Sesbanieae
Cicereae
Vicieae Trifolieae
Loteae
Psoraleeae
Desmodieae
Phaseoleae
Amorpheae
M.3 M.4
M.5
C2.D
P.1 P.2
P.4 P.29
P.30
P.25
P.24 P.23 P.27
P.5
P.22
P.7 P.15
P.18
P.16
P.19 P.20
P.8 P.14
P.8
P.21
P.12
P.6
P.9 P.11
Sophoreae
P.10 Indigofereae
P.13
O
OH O=C
HO
H3COO
CH2
CH3CONH
O O-CONH2
HO
CH3
O
CH3
CH2-OH
N
O O
O O
3 (C16:0 or C16:1 or C14:1 )
O
HO3S-CH3CONH O=C
O=C
o o o o o
CH3-O
H3C HO
H2NCOO
CH2
Current Opinion in Plant Biology
Relationships amongst tribes of the Leguminosae, and the ability of
Rhizobiumsp. NGR234 and R. frediiUSDA257 to nodulate them. All substituents found in the complete family of NodNGR factors are shown near the bottom of the figure, and those structural requirements which NGR234/USDA257 lack, but which seem to be necessary for nodulation
devoid of indigenous rhizobia, but which had been inoculat-ed with a single M. lotiisolate [12]. All contained an identical 500 kb ‘symbiotic island’ integrated into the chromosomal phenyl-alanine transfer RNA gene (phe-tRNA gene) [13•].
At the present moment, the exact nature of the soil bacteria (‘the mezorhizobial recipients’) which have aquired these ‘symbiosis islands’ is not clear. Nevertheless, their aquisition converts formerly saprophytic bacteria into symbionts. At 536 kb, pNGR234a, the symbiotic plasmid of the broad host-range Rhizobiumsp. NGR234, is about the same size as the ‘symbiosis island’, and it confers the ability to nodulate on Agrobacterium tumefaciens. Sequencing of this plasmid highlighted that pNGR234a is particularly interesting, not only because it is a repository of symbiotic promiscuity, but also because of its mosaic genetic structure with origins of replication and transfer very similar to those of Agrobacterium species [14]. These features not only suggest a common ori-gin for symbiotic replicons and tumour-inducing-plasmids, but also exemplify the combinatorial possibilities offered by lateral transfer of genetic information.
Genetic exchange also occurs between symbionts in the soil [15,16]. Once acquired, amplification of the newly inherited genetic information may generate pools of related strains, some of which are better adapted to particular con-ditions than others [17]. Abundant and highly conserved repeats found throughout rhizobial genomes are the sites of amplification. When formed these ‘amplicons’ can be translocated to other plasmids or the chromosome, where they are more stably maintained. Thus, in populations of soil bacteria, natural genetic mechanisms exist which are capable of transforming isolates with widely different chro-mosomal backgrounds into pools of nodulating bacteria.
Nod-factors and host-specificity
What are the bacterial determinants that specify host-range? It has been clear for almost 100 years that rhizobial secretions provoke Had. Some purified Nod-factors pro-voke the extreme curling of the root-hairs (shepherd’s crook formation or Hac), which is the most specific of the morphological responses induced by rhizobia (see [18]). They also induce cortical cell division [19], and permit rhi-zobia to penetrate host-tissues [20,21•]. Since rhizobia that
are unable to synthesise Nod-factors cannot nodulate (i.e. Nod–), these signal molecules are obviously essential
to the symbiosis (see also A Hirsch, this issue pp 320–326). It is thus possible that structural variations in Nod-factors are the determinants of host-specificity. Indeed, mutation of nodH,which encodes a sulphotransferase, leads to the absence of the sulphate group from R. melilotiLCOs, and loss of nodulation of its homologous host M. sativa. Yet, these NodH–mutants are now able to nodulate the non-host V.sativa[22]. Interestingly, almost the only legumes that NGR234 is unable to nodulate are the temperate, herbaceous genera Cicereae, Trifolieae, and Vicieae (Figure 1). This suggests that in evolutionary terms, nodu-lation of this group seems to require a shift away from
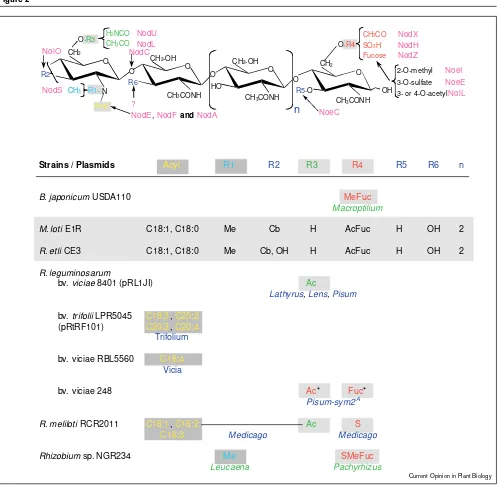
acids to α-β-unsaturated fatty acids and unmodified termini [23] (Figure 1). It should be noted, however, that although nodFEmutants of R. melilotisecrete Nod-factors in which the C16 unsaturated fatty acids are replaced by vaccenic acid, the mutants still form nodules on various Medicago cultivars [24]. Even though a few other correlations between Nod-factor substituents and host-range have been reported (Figure 2), other examples contradict the dogma that Nod-factors determine host-specificity. For instance, Nod-factors of heterologous rhizobia (i.e. isolated from a different legume and unable to nodulate the plant tested) also provoke Hac on root-hairs of Macroptilium atropurpureum[25]. Despite the fact that the Nod-factors produced by R. etliand M. lotiare identical, the two species have distinct host ranges (Phaseolus sp. and Lotus sp. respectively) [26]. Similarly, the major Nod-factors secret-ed by R. leguminosarumbv. trifoliiare identical to two of the predominant Nod-factors produced by R. leguminosarum bv. viciae[27,28], yet these two bacteria also have distinct host ranges. In contrast, two rhizobia that nodulate the same plant may secrete different Nod-factors: R. tropici and R. etliproduce sulphated and acetylfucosylated Nod-factors respectively, but both effectively nodulate Phaseolus vulgaris[29,30]. All symbionts of Glycine maxproduce Nod-factors fucosylated by NodZ, yet null mutations in nodZ have no discernible effect on nodulation of this and many other hosts [31,32]. Thus, although Nod-factors are essen-tial to nodulation and their modification may contribute to host-specificity, this family of signal molecules is probably just one of several elements specifying host range.
NodD proteins and plant exudates
to be responsible for the observed phenotypes. Thus NodD of narrow host-range-rhizobia such as R. meliloti, R. leguminosarum bv. viciae, and R. leguminosarum bv. tri-folii responds to few flavonoids, whereas NodD1 of the broad host-range NGR234 reacts with a large spectrum
of inducing compounds. These include simple phenolic substances such as vanillin and iso-vanillin but also com-pounds that have been identified as inhibitors in other rhizobia. Yet again, these correlations alone are insuffi-cient to explain host-specificity.
Figure 2
Strains / Plasmids Acyl R1 R2 R3 R4 R5 R6 n
B. japonicum USDA110 MeFuc
Macroptilium
M. loti E1R C18:1, C18:0 Me Cb H AcFuc H OH 2
R. etli CE3 C18:1, C18:0 Me Cb, OH H AcFuc H OH 2
R. leguminosarum
bv. viciae 8401 (pRL1JI) Ac
Lathyrus, Lens, Pisum
bv. trifolii LPR5045 C18:3, C20:2 (pRtRF101) C20:3, C20:4
Trifolium
bv. viciae RBL5560 C18:4 Vicia
bv. viciae 248 Ac∗ Fuc∗
Pisum-sym2A
R. meliloti RCR2011 C16:1, C16:2 Ac S
C16:3 Medicago Medicago
Rhizobium sp. NGR234 Me SMeFuc
Leucaena Pachyrhizus
Ac yl
CH2 CH2
HO
O-R4
N O
O
n
O
CH3CONH CH3CONH
O O
OH
R5-O O-R3
CH2-OH O
CH3CONH
CH2-OH O
R1 -R2
R6 NodC
NodE, NodF and NodA NolO
H2NCO NodU
?
NoeC NodS CCH3H3
CH3CO NodX
SO3H NodH
Fucose NodZ
2-O-methyl NoeI
3-O-sulfate NoeE
3- or 4-O-acetylNolL CH3CO NodL
Current Opinion in Plant Biology
Structures of Nod-factors and substituents which alter host-range. Generalised Nod-factor structures are presented in the upper part of the figure, and the table below shows those substituents (shaded) which are necessary for nodulation of the legumes marked in colour. Only those substituents whose presence is essential for nodulation of the genera immediately below are listed. With the exception of the Nod-factors of Mesorhizobium lotiand R. etli, those substituents that have not been shown to play a role in host-specificity are not listed. Although M. lotiand R. etlisecrete identical Nod-factors (boxed), their host-ranges are different. For example, M. lotistrain E1R nodulates
Lotus corniculatus but not Phaseolus vulgaris, whereas R. etlistrain CE3 is Nod+on P. vulgarisand Nod–on L. corniculatus (Cárdenaset
al. [26]). If either strain harbours a flavonoid independent nodDgene, it can nodulate the heterologous host (see text). *The absence of the acetyl group in Nod-factors produced by nodX–mutants can be
phenotypically complemented by the addition of a fucose group (nodZ
encodes a fucosyltransferase)(Ovtsyna et al. [48•]). Ac, acetyl; Cb, carbamoyl; Fuc, fucosyl; H, hydrogen; Me, methyl; S, sulphate; MeFuc, methylfucose; AcFuc, acetylated fucose;
Perhaps the fact that flavonoid–NodD complexes activate nod-gene expression and trigger a chain of events which lead to the secretion of Nod-factors is not enough to account for nodulation of some legumes. Possibly the composition of the Nod-factor mixtures and the levels of Nod-factors in the rhizosphere are also important. USDA257 not only produces a less complicated subset of the NGR234 factors, but it secretes only one-fortieth as many [33]. Amongst the struc-tural differences between the two sets of factors is the absence of an N-methyl group on the non-reducing termi-nus of NodRffactors [9•]. Mutation of nodS, the gene that
encodes the N-methyltranferase deprives NGR234 of the ability to nodulate tropical trees of the genus Leucaena. Conjugation of the nod-box::nodS of NGR234 into USDA257 leads to a twenty-two fold increase in the amount of NodRf factors secreted by USDA257. Due to the N -methyltransferase encoded by nodS[35], these factors are also N-methylated, and the transconjugant gains the ability to nodulate Leuceanaspp. It is not possible to say, however, whether this host-range extension is due to the presence of the N-methyl group or to the elevated Nod-factor levels, although the latter seems more likely given the fact that some micro-symbionts of L. leucocephalado not possess nodS homologues [36].
Co-operation amongst Nod-factors and related signal mole-cules has also been demonstrated. Minami et al. [37,38] examined the effects of LCOs on changes in the expression of early nodulin genes. Many plant genes are up-regulated during nodule development (nodulins). Amongst them are early nodulin genes, which are activated soon after rhizobia enter the plant. One such gene, SrEnod2, encodes a hydroxyproline-rich cell-wall protein, that is expressed in the parenchyma cells of Sesbania rostrata nodules [39•].
Using expression of GmEnod2in G. soyaroots as a marker of nodule initiation, Minami et al. showed that any combina-tion of one active Nod-factor with a non-specific LCO was sufficient to induce GmEnod2 mRNA expression. In con-trast, both synthetic Nod-factors and the chitin pentamer PACT, which is unable to provoke Had on G. soja, induced the transient accumulation of GmEnod40 mRNA. It would thus seem that not only are absolute levels of Nod-factors important for the induction of different components of the nodulation pathway, but also the composition and relative proportions of the mixtures excreted by rhizobia.
Other keys to the legume doors
It seems fairly clear from the above that combinations of flavonoids/NodD proteins/Nod-factors are insufficient to explain host-specificity. Additional factors must, therefore, help control symbiotic development. As Nod-factors permit rhizobia to enter their hosts [20,21•], these additional factors
probably act within the plant (e.g. inside infection threads). Early work showed that the relationship between the num-bers of infection-threads initiated and nodules which develop from them varies from 1.5% to almost 100%, sug-gesting that abortion of inappropriate infections helps control
dence suggests that there is a fine line between symbiosis and pathogenicity, as successful micro-symbionts must also either evade or neutralise the plant defence systems. Specific constituents of the rhizobial cell wall are required for infec-tion-thread initiation and elongation [40•]. These
constituents in R. melilotiinclude polymers of succinoglycan (EPS I) or galactoglucan (EPS II), and capsular polysaccha-rides containing 3-deoxy-D-manno-2-octulosonic acid, (see [18]). Infection threads are composed of plant material, although rhizobia may contribute to the matrix material found inside the threads [41]. Nevertheless null mutations in several genes involved in synthesis of EPS have no effect on nodulation and nitrogen fixation in certain plant/bacteria combinations, suggesting that other elements may substitute for these deficiencies [18]. One possibility is that proteins exported by the type three secretion system (TTSS) may be synergistic with or complementary to extra-cellular polysac-charides in certain symbioses [42•]. In any event, the
principal remains the same — cell wall components and pos-sibly secreted proteins of rhizobia are required for infection thread formation and nodule development. Whether these carbohydrates/proteins play regulatory or structural roles remains to be determined, but it is interesting to note that the rhizobia which produce Nod-factors containing α-β -polyunsaturated fatty acids apparently required for nodulation of temperate herbaceous legumes, also seem to lack the TTSS pathway [42•]. Type I dependent secreted
proteins may also contribute to host-specificity. NodO of R. leguminasarum bv. viciaeallows a nodEmutant of R. legumi-nosarumbv. trifoliito nodulate the non-host V. hirsuta[43], whereas nodOof Rhizobiumsp. BR816 functions as a nodula-tion determinant for L. leucocephala, P. vulgarisand T. repensin diverse rhizobia [44•]. Thus, the existence of two types of
secretion systems, as well as the growing list of known sym-biotically active substances secreted by rhizobia, suggests that the molecular dialogue continues throughout nodule development. Furthermore, the very heterogeneity of secreted substances (acylated and non-acylated oligo- and poly-saccharides, proteins, hormones etc) seems to preclude unique roles for the individual compounds in host-specifici-ty. Rather, we propose that some of these substances complement an important symbiotic function in certain plants, that in other hosts they could trigger defence respons-es, whereas in a third group they are unnecessary because they are either not sensed by the plant or because function-al equivfunction-alents function-already exist in the host. In such a scenario, the secreted substances could play both structural (e.g. in infec-tion-thread development) and signaling roles.
Conclusions
confirms that horizontal transfer of symbiotic information fre-quently occurs in the soil. In this respect, interesting parallels can be drawn to pathogenicity islands (Pais) of virulent bac-teria (see [45]). Both carry either many virulence or nod–fix–nifgenes, are relatively large (up to several 100 kb), present indications of lateral transfer (G and C content and codon-usage differ significantly from the rest of the genome), and are prone to genomic rearrangements. Thus, lateral transfer of symbiotic (or pathogenic) functions followed by adaptation to the new ecological niche probably drives evo-lution of many plant-interacting bacteria. If this is true, strict correlations between rhizobial phylogeny, Nod-factors and host-specificity will remain illusory.
Although it is possible that some major modulator of the interaction between rhizobia and its hosts has been over-looked, it seems more likely that host-specificity is controlled at various levels. The importance of each indi-vidual step would depend on the specific legume–Rhizobium combination and would include levels of Nod-factors in the rhizosphere (and, therefore, of their stability), composition of the families of Nod-factors excreted, as well as post-entry abortion of infection threads triggered by inappropriate extra-cellular polysaccharides or proteins. In any event, a sequence of reactions of variable specificity begins with dif-ferent spectra of phenolic compounds in the rhizosphere of the various hosts. Some of these may or may not interact with NodD protein(s), that in turn bind to quite variable nod-boxes [46,47•], so activating the transcription of nod
-genes which produce varying amounts of different Nod-factor families that may or may not be stable, but which help control rhizobial entry into root-hairs. There, they may provoke abortion, depending on the nature of the extra-cellular carbohydrates or proteins. This sequence of events provides ample flexibility to explain all the nodula-tion patterns observed.
Note added in proof
Two relevant papers on lateral transfer have recently been published [49•,50•].
Acknowledgements
We would like to thank J Dénarié, JA Downie, M Holsters, E Martínez-Romero, R Palacious, CW Ronson, HP Spaink, and G Stacey for helpful comments and ideas during preparation of the manuscript. Dora Gerber gave general support and financial assistance was provided by the Swiss National Fond for Scientific Research (Grant # 31-45921.95).
References and recommended reading
Papers of particular interest, published within the annual period of review, have been annotated.
1. Beyerinck MW: Künstliche Infection von Vicia Fabamit Bacillus radicicola. Ernährungsbedingungen dieser Bacterie.Botanische Zeitung1890, 52:837-843. [Title translation: Artificial infection of Vicia Fabawith Bacillus radicicola. Culture conditions of these bacteria.]
2. Prazmowski A: Die Wurzelknöllchen der Erbse.Landwirtschaftlichen Versuchs-Stationen, Berlin1890, 37:161-238. [Title translation: Root nodules of peas.]
3. Hiltner L: Über die Ursachen, welche die Grösse, Zahl, Stellung und Wirkung der Wurzelknöllchen der Leguminosen bedingen.
Arbeiten aus der Biologischen Abteilung für Land- und
Forstwirthschaft am Kaiserlichen Gesundheitsamte, Berlin1900,
1:177-222. [Title translation: The causes which determine the size, number, position and effect of root nodules on legumes.]
4. Lerouge P, Roche P, Faucher C, Maillet F, Truchet G, Promé J-C, Dénarié J: Symbiotic host-specificity ofRhizobium melilotiis determined by a sulphated and acylated glucosamine oligosaccharide signal.Nature1990, 344:781-784.
5. Spaink HP, Sheeley DM, van Brussel AAN, Glushka J, York WS, Tak T, Geiger O, Kennedy EP, Reinhold VN, Lugtenberg BJJ: A novel highly unsaturated fatty acid moiety of lipo-oligosaccharide signals determines host-specificity of Rhizobium.Nature1991, 354:125-130.
6. Polhill RM: Classification of the Leguminosae.In Phytochemical Dictionary of the Leguminosae, vol. 1, Plants and their Constituents. Edited by Bisby FA, Buckingham J, and Harborne JB. London: Chapman and Hall; 1994: xvi-xxxvii.
7. Hiltner L, Störmer K: Neue Untersuchungen über die
Wurzelknöllchen der Leguminosen und deren Erreger.Arbeiten aus der Biologischen Abteilung für Land- und Forstwirthschaft, am Kaiserlichen Gesundheitsamte (Berlin)1903, 3:151-307. [Title translation: New investigations into the root nodules of the Leguminosae and their causitive agents.]
8. Wilson JK: Leguminous plants and their associated organisms.
Cornell Univ Agric Exp Station, Memoir 221. Cornell University Press; 1939.
9. Pueppke SG, Broughton WJ: Rhizobiumsp. strain NGR234 and
• R. frediiUSDA257 share exceptionally broad, nested host-ranges.
Mol Plant–Microbe Interact1999, 12:293-318.
Nodulation capacities of two promiscuous rhizobia were tested on 452 species of legumes. Correlations between Nod-factor structure and the abil-ity to nodulate, the geographical origins of the legumes, their growth habit, etc were not found.
10. Lewin A, Rosenberg C, Meyer zA H, Wong CH, Nelson L, Manen J-F, Stanley J, Dowling DN, Dénarié J, Broughton WJ: Multiple host-specificity loci of the broad host range Rhizobiumsp. NGR234 selected using the widely compatible legume Vigna unguiculata.
Plant Mol Biol1987, 8:447-459.
11. Martínéz-Romero E, Caballero-Mellado J: Rhizobiumphylogenies and bacterial genetic diversity.Critical Rev Plant Sci 1996, 15:113-140.
12. Sullivan JT, Patrick HN, Lowther WL, Scott DB, Ronson CW:
Nodulating strains of Rhizobium lotiarise through chromosomal symbiotic gene transfer in the environment.Proc Natl Acad Sci USA1995, 92:8985-8989.
13. Sullivan JT, Ronson CW: Evolution of rhizobia by acquisition of a
• 500-kb symbiosis island that integrates into a phe-tRNA gene.
Proc Natl Acad Sci USA1998, 95:5145-5149.
Convincing evidence that a mobile 500 kb ‘symbiosis island’ is transferred to associated rhizospheric saprophytes rendering them symbiotic.
14. Freiberg C, Fellay R, Bairoch A, Broughton WJ, Rosenthal A, Perret X:
Molecular basis of symbiosis between Rhizobium and legumes.
Nature 1997, 387:394-401.
15. Broughton WJ, Samrey U, Stanley J: Ecological genetics of Rhizobium meliloti: symbiotic plasmid transfer in the Medicago sativarhizosphere.FEMS Microbiol Lett1987, 40:251-255.
16. Schofield PR, Gibson, AH, Dudman WF, Watson JM: Evidence for genetic exchange and recombination of Rhizobium symbiotic plasmids in a soil population.Appl Environ Microbiol. 1987,
53:2942-2947.
17. Mavingui P, Flores M, Romero D, Martínez-Romero E, Palacios R:
Generation of Rhizobiumstrains with improved symbiotic properties by random DNA amplification (RDA).Nat Biotech1997,
15:564-569.
18. Perret X, Staehelin C, Broughton WJ:Molecular basis of symbiotic promiscuity.Microbiol Mol Biol Rev1999, in press.
19. Truchet G, Roche P, Lerouge P, Vasse J, Camut S, de Billy F, Promé J-C, Dénarié J: Sulphated lipo-oligosaccharide signals of Rhizobium melilotielicit root nodule organogenesis in alfalfa.
Nature 1991, 351:670-673.
20. Relic´ B, Perret X, Estrada-Garcia MT, Kopcinska J, Golinowski W, Krishnan HB, Pueppke SG, Broughton WJ: Nod-factors of Rhizobium are a key to the legume door.Mol Microbiol1994, 13:171-178.
21. D’Haeze W, Gao M-S, De Rycke R, Van Montagu M, Engler G,
22. Roche P, Debellé F, Maillet F, Lerouge P, Faucher C, Truchet G, Dénarié J, Promé J-C: Molecular basis of symbiotic host specificity in Rhizobium meliloti: nodHand nodPQgenes encode the sulfation of lipochito-oligosaccharide signals.Cell1991, 67:1131-1143.
23. Yang GP, Debellé F, Ferro M, Maillet F, Schiltz O, Vialas C, Savagnac A, Promé J-C, Dénarié J:RhizobiumNod factor structure and the phylogeny of temperate legumes.In Biological Nitrogen Fixation for the 21stCentury. Edited by Elmerich C, Kondorosi A and Newton WE. Kluwer Academic Publishers, Dordrecht; 1998:185-188.
24. Ardourel M, Demont N, Debellé F, Maillet F, de Billy F, Promé J-C, Dénarié J, Truchet G: Rhizobium melilotilipooligosaccharide nodulation factors: different structural requirements for bacterial entry into target root hair cells and induction of plant symbiotic developmental responses.Plant Cell1994, 6:1357-1374.
25. Relic´ B, Talmont F, Kopcinska J, Golinowski W, Promé J-C, Broughton WJ: Biological activity of Rhizobiumsp. NGR234 Nod-factors on Macroptilium atropurpureum.Mol Plant–Microbe Interact 1993, 6:764-774.
26. Cárdenas L, Dominguez J, Quinto C, Lopez-lara IM, Lugtenberg BJJ, Spánk HP, Rademaker GJ, Haverkamp J, Thomas-Oates JE:Isolation, chemical structure and biological activity of the lipo-chitin oligosaccharide nodulation signals from Rhizobium etli.Plant Mol Biol1995, 29:453-464.
27. Spaink HP, Bloemberg GV, van Brussel AAN, Lugtenberg BJJ, van der Drift KMGM, Haverkamp J, Thomas-Oates JE: Host specificity of Rhizobium leguminosarumis determined by the hydrophobicity of highly unsaturated fatty acyl moieties of the nodulation factors.
Mol Plant–Microbe Interact1995, 8:155-164.
28. Orgambide GG, Lee JI, Hollingsworth RI, Dazzo FB: Structurally diverse chitolipooligosaccharide Nod factors accumulate primarily in membranes of wild-type Rhizobium leguminosarum biovar trifolii.Biochemistry1995, 34:3832-3840.
29. Poupot R, Martínez-Romero E, Promé J-C: Nodulation factors from Rhizobium tropiciare sulfated or non-sulfated
chitopentasaccharides containing an N-methyl-N-acylglucosamine terminus.Biochemistry1993, 32:10430-10435.
30. Poupot R, Martínez-Romero E, Gautier N, Promé J-C: Wild-type Rhizobium etli, a bean symbiont, produces acetyl-fucosylated, N -methylated, and carbamoylated nodulation factors.J Biol Chem 1995, 270:6050-6055.
31. Stacey G, Luka S, Sanjuan J, Banfalvi Z, Nieuwkoop AJ, Chun JY, Forsherg LS, Carlson R: nodZa unique host-specific nodulation gene, is involved in the fucosylation of the lipooligosaccharide nodulation signal from Bradyrhizobium japonicum.J Bacteriol 1994, 176:620-633.
32. Quesada-Vincens D, Fellay R, Nasim T, Viprey V, Burger U, Promé J-C, Broughton WJ, Jabbouri S:Rhizobiumsp. NGR234 NodZ protein is a fucosyltransferase.J Bacteriol1997, 179:5087-5093.
33. Relic´ B, Staehelin C, Fellay R, Jabbouri S, Boller T, Broughton WJ: Do Nod-factor levels play a role in host-specificity?In Proceedings of the First European Nitrogen Fixation Conference. Edited by Kiss GB and Endre G. Officina Press: Szeged, Hungary; 1994:69-75.
34. Spaink HP, Wijffelman CA, Pees E, Okker RJH, Lugtenberg BJJ:
Rhizobiumnodulation gene nodDas a determinant of host specificity.Nature1987, 328:337-340.
35. Jabbouri S, Fellay R, Talmont F, Kamalaprija P, Burger U, Relic´ B, Promé J-C, Broughton WJ: Involvement of nodSin N-methylation and nodUin 6-O-carbamoylation of Rhizobium sp. NGR234 Nod factors.J Biol Chem1995, 270:22968-22973.
36. Krishnan HB, Lewin A, Fellay R, Broughton WJ, Pueppke SG:
Differential expression of nodSaccounts for the varied abilities of Rhizobium frediiUSDA 257 and Rhizobiumsp. NGR234 to nodulate Leucaenaspp.Mol Microbiol1992, 6:3321-3330.
37. Minami E, Kouchi H, Carlson RW, Cohn JR, Kolli VK, Day RB, Ogawa T, Stacey G: Cooperative action of lipo-chitin nodulation signals on the induction of the early nodulin, ENOD2 in soybean roots.Mol Plant–Microbe Interact1996, 9:574-583.
38. Minami E, Kouchi H, Cohn JR, Ogawa T, Stacey G:Expression of the early nodulin, ENOD40, in soybean roots in response to various lipo-chitin signal molecules.Plant J1996, 10:23-32.
The 3′untranslated region of SrEnodis required for its expression in the parenchyma cell layer of the nodules. Expression of SrEnod2is inducible by cytokinins, adding plant hormones to the list of substances known to modu-late expression of this gene (see [38]). Thus Nod-factors and cytokinins joint-ly modulate the activity of some of the same components of the signal response pathway.
40. Cheng H-P, Walker GC: Succinoglycan is required for initiation and
• elongation of infection threads during nodulation of alfalfa by Rhizobium meliloti.J Bacteriol1998, 180:5183-5191.
Shows clearly that individual exo-genes, which are involved in the synthesis of succinoglycan, affect infection-thread development.
41. Kijne JW: The Rhizobiuminfection process.In Biological Nitrogen Fixation. Edited by Stacey G, Burris RH, and Evans HJ: Chapman and Hall; New York; 1992:349-398.
42. Viprey V, Del Greco A, Golinowski W, Broughton WJ, Perret X:
• Symbiotic implications of type III protein secretion machinery in Rhizobium.Mol Microbiol1998, 28:1381-1389.
Sequencing of pNGR234a revealed a gene cluster which encodes a flavonoid inducible type III protein secretion system. Builds on data which showed that protein secretion plays a role in host-specificity.
43. Economau A, Davies AE, Johnston AWB, Downie JA: The Rhizobium leguminosarum biovar viciae nod0 gene can enable a nodE mutant of Rhizobium leguminosarum biovar trifolií to nodulate vetch. Microbiology1994, 140:2341-2347.
44. Vlassak KM, Luyten E, Verreth C, van Rhijn P, Bisseling T,
• Vanderleyden J: The Rhizobiumsp. BR816 nodOgene can function as a determinant for nodulation of Leucaena leucocephaia, Phaseolus vulgarisand Trifolium repensby a diversity of Rhizobium spp.Mol Plant–Microbe Interact1998, 11:383-392. Shows that nodO is not confined to strains that nodulate Cicereae, Trifolieae, and Vicerea, but is also present in rhizobia that associate with tropical legumes. Functionally complements mutants in nodSof NGR234 for nodulation of L. leucoceplaia.
45. Hacker J, Blum Oehier G, Mühldorfer I, Tschäpe H: Pathogenicity islands of virulent bacteria: structure, function and impact on microbial evolution.Mol Microb1997, 23:1089-1097.
46. van Rhijn P, Vanderleyden J: The Rhizobium-plant symbiosis.
Microbiol Rev1995, 59:124-142.
47. Perret X, Freiberg C, Rosenthal A, Broughton WJ, Fellay R:
High-• resolution transcriptional analysis of the symbiotic replicon of Rhizobium sp. NGR234.Mol Microbiol1999, 32:415-425.
pNGR234awas divided into 441 segments representing all coding and inter-genic regions. Each segment was amplified by PCR, transferred to filters and probed with radioactively labelled-RNA. Genes involved in the synthesis of Nod-factors were induced rapidly after the addition of flavonoids, while others thought to act within the plant (e.g. those encoding the type three secretion machine) responded more slowly. Many more transcripts were found in bac-teroids of determinate- as opposed to indeterminate-nodules.
48. Ovtsyna AO, Geurts R, Bisseling T, Lugtenberg BJJ, Tikhonovich IA,
• Spaink H: Restriction of host range by the sym2allele of Afghan pea is nonspecific for the type of modification at the reducing terminus of nodulation signals.Mol Plant–Microbe Interact1998,
11:418-422.
A clear demonstration that the exact nature of Nod-factor substitutents is less important than their position. Nodulation of the primitive P. sativum cultivar Afghanistan was thought to require a 6-O-acetate group on the reducing ter-minus (added by NodX), but it can be functionally replaced by a 6-O-fucose group (nodZencodes a fucosyltransferase).
49. Karaolis DKR, Somara S, Maneval DR, Johnson JA, Kaper JB: A
• bacteriophage encoding a pathogenicity island, a type-IV pilus and a phage receptor in cholera bacteria. Nature1999, 399:375-379. Many bacteria carry pathogenicity islands — DNA sequences that encode viru-lence related proteins. These include specialised molecular machines to inject-bacterial products into their animal or plant hosts, molecules involved in signal transduction and toxins. Here the authors demonstrate an interplay between two pathogenicity islands of the bacterium Vibrio choleraeand its bacterio-phage CTXψ, which carries the cholera toxin genes.
50. Nelson KE, Clayton RA, Gill SR, Gwinn ML, Dodson RJ, Haft DH,