Differential expression of plastidic aldolase genes in
Nicotiana
plants under salt stress
Shigehiro Yamada *, Toshiyuki Komori, Akiko Hashimoto, Shigeru Kuwata,
Hidemasa Imaseki, Tomoaki Kubo
Plant Breeding and Genetics Research Laboratory,Japan Tobacco Inc., 700Higashibara,Toyoda-cho,Iwata-gun,Shizuoka438-0802,Japan
Received 18 May 1999; received in revised form 14 December 1999; accepted 20 December 1999
Abstract
Two homologous genes of plastidic fructose-1,6-bisphosphate aldolase (AldP) isozymes were isolated from green leaves of a salt stress-tolerant Nicotiana species, Nicotiana paniculata, by differential screening. The products of the corresponding genes,
NpAldP1 and NpAldP2, were 91% identical to each other and 70 – 85% identical to the other known plant plastidic aldolases. Although these two genes showed similar organ-specific expression and daily cycles, their responses to salt stress differed: mRNA accumulation ofNpAldP2 increased, but that ofNpAldP1 slightly decreased. The mRNA accumulations of their counterparts of two otherNicotianaspecies,NeAldP1 and NeAldP2 (Nicotiana excelsior), andNaAldP1 andNaAldP2 (Nicotiana arentsii) were studied under the same stress condition.N.arentsiiconserved accumulation profiles similar toN.paniculata, butN.excelsiordid not. InN.excelsior, accumulation of NeAldP1 decreased to 50% of the control after stress and gradually recovered thereafter, whereas accumulation ofNeAldP2 temporarily decreased and reached 250% of the control by the third day of stress. Southern blot analysis indicated thatNpAldP1, NpAldP2,NaAldP1, and NaAldP2 include one or two closely related genes andNeAldP1 andNeAldP2 several. © 2000 Elsevier Science Ireland Ltd. All rights reserved.
Keywords:Nicotiana; Salt stress; Aldolase
www.elsevier.com/locate/plantsci
1. Introduction
Environmental stresses often strongly affect crop production, and improvement in plant ability to tolerate such stresses is one of the most impor-tant objectives in plant science. Water-deficit stress, which is mainly due to high salinity and/or drought, is one of the most common stresses. Several groups have achieved this mainly by intro-ducing foreign genes into host plants to enhance osmoprotectant productions; however, the effects were limited [1 – 4]. Therefore, introduction of mul-tiple mechanisms is assumed essential to confer
extensive stress tolerance. To find novel stress-tol-erance mechanisms, the authors have been investi-gating the response of Nicotiana species to water-deficit stress. Plants in this genus are not classified as halophytic plants, but some of their native habitats are harsh, either dry or salty. A total of 57 Nicotiana species were screened for water-deficiency tolerance, and Nicotiana panicu
-lata and Nicotiana excelsior were found to be water-deficit stress-tolerant. These species can grow as well as unstressed plants even under the salt stress imposed by 250 mM NaCl (Komori, submitted). The Nicotiana species also contain stress-sensitive species, such as Nicotiana arentsii. This species shows slow growth under the same conditions (Komori, submitted). To elucidate stress-tolerance mechanisms of these species using the molecular approach, differential screening was * Corresponding author. Tel.: +81-538-327111; fax: +
81-538-336046.
E-mail address:[email protected] (S. Yamada)
attempted and it succeeded in isolating stress-in-ducible genes from those species. Among the cD-NAs obtained from the green leaves of N.
paniculata, two clones were found with a homol-ogy to plastidic fructose-1,6-bisphospate aldolase (AldP) genes.
AldP catalyzes the cleavage of fructose-1,6-bis-phosphate (FBP) into D
-glyceraldehyde-3-phos-phate (GAP) and dihydroxyacetone phos-glyceraldehyde-3-phos-phate (DHAP). Two forms of aldolase are present in higher plants: cytoplasmic and plastidic. The cy-toplasmic aldolase (AldC) gene is inducible un-der anaerobic conditions and is consiun-dered to have an important role in producing ATP by stimulating glycolysis under such conditions [5 – 9]. AldP is involved in the photosynthetic carbon reduction cycle and catalyzes the synthesis of FBP from GAP and DHAP. AldP gene expres-sion is regulated by light [10], but the effect of other environmental factors on AldP is nu-known. Here, the isolation of two AldP genes from a salt-tolerant Nicotiana species, N. panicu
-lata is reported and their expression regulation among three Nicotiana species are compared with different salt-tolerance abilities under salt stress.
2. Materials and methods
2.1. Plant materials
Plants were grown from seeds in a greenhouse. Seedlings were transferred to plastic pots (10 cm in diameter) filled with a mixture [1:1 (v/v)] of vermiculite and Hydroball (small particles of brick, Jongkind Ground B.V., Aalsmeer, the Netherlands) 3 weeks after germination. The plants were watered daily with 100 ml of 25% Hoagland’s solution (Hoagland’s Solution No. 2, Sigma). They were grown in the greenhouse for an additional 7 days, and then were transferred to a growth chamber 1 week before stress was applied. The growth chamber was operated on a 12-h light (23°C)/12-h dark (20°C) cycle. Light was provided by 400 W metal halide lamps at a constant 500 – 600 mEm−2 s−1. Salt stress was initiated 1 h after illumination had started by watering with 100 ml of 25% Hoagland’s solu-tion containing 250 mM NaCl and 100 ml of the same solution was watered daily thereafter.
Con-trol plants were watered daily with 100 ml of 25% Hoagland’s solution only. Plants for the ex-periment examining organ specificity were grown in the greenhouse from sowing to anthesis. Plants for the experiment examining the daily cycle were grown in the growth chamber under the same conditions as above.
3. Differential screening
Total RNA was isolated from green leaves of
N. paniculata 24 h after stress was applied, as described by Ostrem et al. [11]. Poly(A)+ RNA
was purified using a QuickPrep mRNA Purifica-tion Kit (Amersham Pharmacia Biotech), and a cDNA library was constructed with the poly(A)+
RNA using a ZAP-cDNA Synthesis Kit (Strata-gene). This library was subjected to a differential screening. Plaque lifts were done as described in the manufacturer’s manual, except that 3000 plaques were plated per plate. Membranes for hybridization were made in duplicate from each plate. 32P-labeled first-strand cDNA probes were prepared from the above poly(A)+ RNA from
stressed plants and poly(A)+ RNA isolated from
control plants. The reverse transcription reaction was done at 37°C for 1 h in 50 ml of reaction mixture containing 50 mM Tris – HCl (pH 8.3), 75 mM KCl, 3 mM MgCl2, 10 mM dithiothrei-tol, 1 mM dATP, 1 mM dGTP, 1 mM dTTP, 100 ng of poly(A)+ RNA, 2.5 mg of random
primers (9 mer) (Takara, Shiga, Japan), 35.5 U of RNAguard (Amersham Pharmacia Biotech), 1.85 MBq of [a-32P]dCTP (Amersham Pharmacia Biotech), and 10 U of M-MLV reverse transcrip-tase (Toyobo, Osaka, Japan). Unincorporated [a -32P]dCTP was removed from the mixture using a
Quick Spin Column (Boehringer Mannehim) af-ter the reaction. Each set of membranes was hy-bridized with either the control probe or the stress probe. The membranes were hybridized at 65°C for 20 h in a solution containing 125 mM Na2HPO4, 1 mM EDTA, and 7% SDS. After hybridization the membranes were washed twice at 65°C for 20 min in a solution containing 2×
3.1. DNA and deduced amino acid sequence analyses
Nucleotide sequences of the clones were deter-mined using an automatic DNA sequencer (Prism 310, PE Applied Biosystems). DNA and deduced amino acid sequence analyses of obtained genes and their products, including phylogenetic study, were made using Genetyx-Mac ver. 9.0 (Software Development, Tokyo, Japan). Database searches were made using the GenomeNet WWW Server of Kyoto University (http://www.genome.ad.jp).
3.2. Northern blot analysis
Total RNA was isolated from the leaves of stressed plants and leaves, roots, flowers, stems, or petioles of unstressed plants, as described by Os-trem et al. [11]. Northern blotting was done with 5
mg of LiCl-purified total RNA, as described by Vernon and Bohnert [12]. 32P-labeled probes were prepared from the whole cDNA regions using a Rediprime DNA labeling system (Amersham Pharmacia Biotech) and were used for hybridiza-tion. Hybridization was done in the same condi-tion as for differential screening. Hybridized membranes were analyzed and the signals were quantified using FUJX BAS1000.
3.3. Southern blot hybridization
Total DNA was isolated from leaves of each examined species, as described by Hiei et al. [13]. Southern blot hybridization was done with 20 mg of total DNA digested with restriction enzymes, using the same method as for Northern blot analysis.
4. Results
4.1. Isolation and sequence analyses of AldP genes
In differential screening of a leaf cDNA library fromN.paniculata, which had been stressed for 24 h by adding 250 mM NaCl to the nutrient solu-tion, 2×105 plaques were screened and 61 inde-pendent clones were isolated. By exploratory partial sequence analyses, two clones were found to have a high homology to aldolases from higher plants. A comparison of the complete nucleotide
sequences of these clones showed that the clones are products of different genes, NpAldP1 and
NpAldP2. The nucleotide sequences for these clones were deposited in the EMBL/GenBank/
DDBJ under accession numbers AB027001 and AB027002. These clones had 1384 and 1431 bp, respectively (excluding poly adenylation sites), and had a reading frame of 395 and 398 amino acids, respectively. They are 75% homologous at the nucleotide level and 91% identical at the amino acid level. An amino acid sequence comparison with known aldolases from higher plants showed a putative transit peptide region at the amino termi-nal end of these products, which led to the conclu-sion that these products were AldP (Fig. 1a). A phylogenetic study based on the amino acid se-quences also supported this conclusion (Fig. 1b). Their similarity with other AldP and AldC was 70 – 85 and 54 – 60%, respectively.
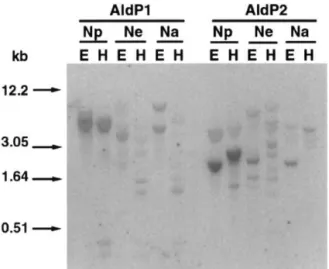
4.2. Gene complexity
Hybridization of NpAldP1 or NpAldP2 to total DNA of N. paniculata, N. excelsior, and N.
arentsii was done under high stringency condi-tions. NpAldP1 cDNA has no EcoRI site and three HindIII sites, and NpAldP2 cDNA has one
EcoRI site and one HindIII sites. Therefore, the band patterns in N.paniculataindicate this species has one or two copies of the AldP1 and AldP2 genes (Fig. 2), but further information is needed about restriction sites of their genomic sequences to give more precise conclusions. There were a few bands in each lane in N. arentsii, and this pattern indicates that this species has one or two copies of AldP1 and AldP2 genes (Fig. 2). The band pat-terns inN. excelsiorwere distinct from those ofN.
paniculata or N. arentsii; there were several bands in each lane (Fig. 2). This result indicates that the gene structure of N. excelsior is distinct from the other two species and has several copies of AldP1 and AldP2 genes.
4.3. Organ specificity of NpAldP1 and NpAldP2
expression
Fig. 2. Southern blot analysis of AldP1 and AldP2 genes. Twenty mg of genomic DNA from N. paniculata (Np), N.
excelsior (Ne), or N. arentsii (Na) was digested with the restriction enzymes (E;EcoRI, H;HindIII) and subjected to agarose gel electrophoresis. DNA was blotted onto nylon membranes and was hybridized with32P-labeled probes.
genes were high in green leaves, low in flowers, stems, and petioles, and undetectable in roots. As expected, high mRNA accumulations were ob-served in organs with a high chloroplast content.
4.4. mRNA Accumulation of AldP genes under salt stress
Time courses of the mRNA levels of AldP genes under salt stress were analyzed by Northern blot analysis in N. paniculata, N. excelsior, and N.
arentsii (Fig. 4a). The signals were quantified, and the stress/control ratios (Fig. 4b) and relative strengths (Fig. 4c) at each time point were plotted. In Fig. 4c, signal strength of the control at 0 h in each blot was taken as unity. N. paniculataand N.
arentsii showed similar expressions of each aldo-lase gene: the amount of AldP1 mRNA gradually decreased, but the accumulation of AldP2 slightly increased under salt stress (Fig. 4b). In contrast, salt stress distinctively affected accumulations of mRNA of AldP genes in N. excelsior: the mRNA level of NeAldP1 decreased to 50% of the control within 3 h and gradually recovered by the fifth day; the mRNA level ofNeAldP2 decreased to less than 50% of the control within 3 h, but it recov-ered within 24 h and exceeded the control level thereafter, reaching 250% of the control (Fig. 4b). Fig. 4c, which shows the relative strength of each signal, also indicates that the mRNA amount of
NeAldP2 was distinctly increased by salt stress. These results showed that the AldP2 gene was up-regulated under salt stress in these species with different profiles, while the AldP1 gene was nega-tively affected by salt stress.
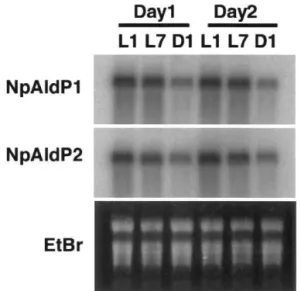
4.5. Daily changes of NpAldP1 and NpAldP2
mRNA accumulation
Fig. 5 shows the daily cycles of NpAldP1 and
NpAldP2 mRNA accumulation in green leaves of unstressed N. paniculata. The mRNA amounts Fig. 3. Northern blot analysis of NpAldP1 andNpAldP2 for
organ specificity. Five mg of LiCl-purified RNA prepared from each organ was resolved on formaldehyde agarose gel and blotted onto nylon membranes. Northern hybridization was done with 32P-labeled probes. L, leaves; R, roots; F,
flowers; S, stems; P, petioles. The bottom panel shows an EtBr-stained gel.
Fig. 1. (a) Alignment of deduced amino acid sequences ofNpAldP1 andNpAldP2 products compared with the deduced amino acid sequences of plastidic (RICCHLALD, SOALDCHL) and cytoplasmic (RICAC1, SOALDCYT) aldolases from higher plants. Asterisks and dots indicate identical and homologous residues, respectively. (b) Phylogenetic tree of plant aldolases. Plastidic and cytoplasmic groups are enclosed in boxes. STPLASALD; Solanum tuberosum (Y10380), PSALDIA; Pisum sati6um (M97476),
PSALDIB;Pisum sati6um(M97477), RICCHLALD;Oryza sati6a(D13513), SOALDCHL;Spinacia oleracea(X66814),
CRALD-CHL;Chlamydomonas reinhardtii(X69969), MZEALD;Zea mays(M16220), OSFBPA;Oryza sati6a(X53130), RICAC1;Oryza
sati6a (D50301), MCAF3124; Mesembryanthemum crystallinum (AF003124), SOALDCYT; Spinacia oleracea (X65742),
Fig. 5. Northern blot analysis of NpAldP1 andNpAldP2 for daily cycles. Total RNA was isolated from green leaves ofN.
paniculataat the indicated time for 2 consecutive days. Five
mg of LiCl-purified RNA was resolved on formaldehyde agarose gel and was blotted onto nylon membranes. Northern hybridization was done with32P-labeled probes. L1, 1 h after
illumination start; L7, 7 h after illumination start; D1, 1 h after illumination termination.
for Southern analysis require 90% homology or higher to hybridize. The band patterns of AldP1 and AldP2 were different from each other in the other two species (Fig. 4), indicating that plastidic aldolase genes can be grouped in two sub-families in these species, too. Although more work is re-quired to determine the exact number of copies of each gene in each species, N. excelsior seems to have more copies than the other two species. N.
paniculata and N. arentsii have 24 chromosomes, whereas N. excelsior has 38 [20]. Therefore the genome structure of N. excelsior is predicted to be different from the other two. This difference in N.
excelsior may reflect the distinct gene dosage. Effects of environmental stress on AldP gene expression have not yet been reported. It was found that AldP2 was up-regulated by salt stress (Fig. 4), which is the first reported instance of environmental stress inducing AldP gene expres-sion. Expression of AldP1 genes was suppressed by salt stress. Although some characteristics, such as amino acid sequences of their products, organ specificity, and daily cycles, are very similar, AldP1 and AldP2 might have different physiologi-cal roles. AldC gene expression is induced under anaerobic conditions to facilitate glycolysis and produce ATP in higher plants [5 – 9]. Because AldC and AldP are involved in different metabolic pathways and catalyze the reaction in opposite directions from each other, the physiological meaning of AldP induction must be different from that of AldC.
One interpretation is that the AldP regulation itself is not involved in the stress tolerance mecha-nisms represented by osmoprotectant production, but that the regulation is derivative. This would be supported by the observation that AldP2 behavior under salt stress seems to correlate with the char-acteristics of each species. The ability to accumu-late dry matter is similar in N. paniculata and N.
arentsii, approximately 60% of the control plants 9 days after stress initiation, although the former is stress tolerant and the latter is stress sensitive. were high under illumination (1 and 7
h-illumina-tion for lanes L1 and L7, respectively) and de-creased in the dark (1 h-dark for lane D1). This result suggests that the decrease in signal 8 h after the stress initiation in the control plants (Fig. 4c) is due to the daily cycles.
5. Discussion
Genes for AldP have been isolated from some higher plants [14,15] and Chlamydomonas[16]. In-vestigation of their genomic structures [10,17 – 19] revealed only one isogene for the AldP gene in the genome of rice, spinach, andChlamydomonas and two isogenes in pea [18]. In this study, the se-quence analyses of cDNA clones and the results of Southern blot analyses proved that two AldP genes,NpAldP1 andNpAldP2, exist in the genome of N. paniculata. Each of these genes has one or two copies in the genome. The conditions we used
Fig. 4. Northern blot analysis of AldP1 and AldP2 genes for stress response. (a) Total RNA was isolated from green leaves of each species at the indicated times after stress initiation. Fivemg of LiCl-purified RNA was resolved on formaldehyde agarose gel and was blotted onto nylon membranes. Northern hybridization was done with32P-labeled probes. (b) The signals were quantified
However, N. excelsior, which is stress tolerant, accumulates dry matter at 80% of the control plant (Komori, submitted). This implies that pho-tosynthetic activity is higher inN.excelsiorthan in the other two species under such conditions. N.
excelsior also shows a characteristic profile in leaf water potential changes under salt stress. It retains a leaf water potential higher than the other two species on the first day of salt application, then the water potential starts to decrease on the second day (Komori, unpublished data). Based on these observations, it was thought that N. paniculata
and N. excelsior have different tolerance mecha-nisms. The tolerance mechanism in N. excelsior
seems to include avoiding unnecessary water losses in the early phase of stress. To achieve this, it might down-regulate photosynthetic activity, and as a derivative result, the expression of both
NeAldP1 and NeAldP2 is down-regulated tempo-rarily. Then, the expression of the two AldP genes recovers when the plant completes its tolerance mechanisms and resumes normal photosynthesis. A recent study using an antisense technique showed that small changes in AldP activity have marked consequences for photosynthesis [21]. It is, therefore, possible that regulating photosynthetic activity at the aldolase step is efficient, and so N.
excelsior down-regulates AldP gene expression un-der salt stress. The increase in NeAldP2 mRNA accumulation may be to compensate for aldolase activity reduced by salt stress. Up-regulation of the AldP2 gene in N. paniculata and N. arentsii is not as clear as that in N. excelsior, and this difference may reflect the photosynthetic ability of these species under stress.
Another interpretation is that the AldP regula-tion is involved in stress tolerance mechanisms. In the antisense study [21], Haake et al. observed an increase in GAP and DHAP caused by a decrease in aldolase expression. If photosynthesis activity is not seriously damaged immediately after the initia-tion of salt stress, GAP and DHAP concentrainitia-tion increases, and these triose phosphates are exported to the cytosol by the triose-phosphate translocator [22] and converted into hexoses. These hexoses could function as osmoprotectants or be used to produce other osmoprotectants. By reducing AldP expression, N. excelsior may change the carbon partitioning transiently, i.e. reducing starch accu-mulation in the chloroplasts and enhancing sugar formation in the cytosol to increase the osmolarity
there. Further analyses of photosynthetic activity, other photosynthesis-involved genes, and sugar ac-cumulation under salt stress are needed to clarify the meaning of AldP gene regulation.
N. excelsiormight have acquired stress tolerance mechanisms by having a stress-responsive aldolase gene, while this might not have occurred in the other two species. Because N. excelsior is thought to have several copies of the AldP2 gene, the increase of AldP2 mRNA level under salt stress could reflect an increase of a specific member of the gene family. A thorough investigation of ge-nomic structure of aldolase genes in these species is needed to clarify this assumption.
Acknowledgements
This work was supported by the Petroleum En-ergy Center of Japan and the Research Associa-tion For Biotechnology. We thank Yoshiko Amma and Mika Tabayashi for their technical assistance.
References
[1] H. Hayashi, Alia, L. Mustardy, P. Deshnium, M. Ida, N. Murata. Transformation ofAarbidopsis thalianawith the
codAgene for choline oxidase; accumulation of glycine-betaine and enhanced tolerance to salt and cold stress. Plant J. 12 (1997) 133 – 142.
[2] P.B.K. Kishor, Z. Hong, G.-H. Miao, C.-A.A. Hu, D.P.S. Verma, Overexpression of D1
-pyrroline-5-car-boxylate synthetase increases proline production and confers osmotolerance in transgenic plants, Plant Phys-iol. 108 (1995) 1387 – 1394.
[3] B. Shen, R.G. Jensen, H.J. Bohnert, Increased resistance to oxidative stress in transgenic plants by targeting man-nitol biosynthesis to chloroplasts, Plant Physiol. 113 (1997) 1177 – 1183.
[4] M.C. Tarczynski, R.G. Jensen, H.J. Bohnert, Stress pro-tection of transgenic tobacco by production of the os-molyte mannitol, Science 259 (1993) 508 – 510.
[5] D.L. Andrews, D.M. MacAlpine, J.R. Johnson, P.M. Kelly, B.G. Cobb, M.C. Drew, Differential induction of mRNAs for the glycolytic and ethanolic fermentative pathways by hypoxia and anoxia in maize seedlings, Plant Physiol. 106 (1994) 1575 – 1582.
[6] C.V. Mujer, M.E. Rumpho, J.-J. Lin, R.A. Kennedy, Constitutive and inducible aerobic and anaerobic stress proteins in the Echinochloa complex and rice, Plant Physiol. 101 (1993) 217 – 226.
[8] M. Umeda, H. Uchimiya, Differential transcript levels of genes associated with glycolysis and alchohol fermenta-tion in rice plants (Oriza sati6a L.) under submergence
stress, Plant Physiol. 106 (1994) 1015 – 1022.
[9] P.M. Kelly, M. Freeling, Anaerobic expression of maize fructose-1,6-diphosphate aldolase, J. Biol. Chem. 259 (1984) 14180 – 14183.
[10] Y. Kagaya, H. Nakamura, S. Hidaka, S. Ejiri, K. Tsut-sumi, The promoter from the rice nuclear gene enoding chloroplast aldolase confers mesophyll-specific and light-regulated expression in transgenic tobacco, Mol. Gen. Genet. 248 (1995) 668 – 674.
[11] J.A. Ostrem, S.W. Olson, J.M. Schmitt, H.J. Bohnert, Salt stress increases the level of translatable mRNA for phosphoenolpyruvate carboxylase in Mesembryanthe
-mum crystallinum, Plant Physiol. 84 (1987) 1270 – 1275. [12] D.M. Vernon, H.J. Bohnert, A novel methyl transferase
induced by osmotic stress in the facultative halophyte
Mesembryanthemum crystallinum, EMBO J. 11 (1992) 2077 – 2085.
[13] Y. Hiei, S. Ohta, T. Komari, T. Kumashiro, Efficient transformation of rice (Oriza sati6a L.) mediated by
Agrobacterium and sequence analysis of the boundaries of the T-DNA, Plant J. 6 (1994) 271 – 282.
[14] S. Chopra, R. Dolferus, M. Jacobs, Cloning and se-quencing of the Arabidopsis aldolase gene, Plant Mol. Biol. 15 (1990) 517 – 520.
[15] B. Pelzer-Reith, A. Penger, C. Schnarrenberger, Plant aldolase: cDNA and deduced amino-acid sequences of the chloroplast and cytosol enzyme from spinach, Plant Mol. Biol. 21 (1993) 331 – 340.
[16] B. Pelzer-Reith, S. Freund, C. Schnarrenberger, H. Yat-suki, K. Hori, The plastid aldolase gene from Chlamy
-domonas reinhardtii: intron/exon organization, evolution, and promoter structure, Mol. Gen. Genet. 248 (1995) 481 – 486.
[17] H. Nakamura, W. Satoh, S. Hidaka, Y. Kagaya, S. Ejiri, K. Tsutsumi, Genomic structure of the rice aldolase isozyme C-1 gene and its regulation through a Ca2+
-me-diated protein kinase-phosphatase pathway, Plant Mol. Biol. 30 (1996) 381 – 385.
[18] K. Razdan, R.L. Heinrikson, H. Zurcher-Neely, P.W. Morris, L.E. Anderson, Chloroplast and cytoplasmic enzymes: isolation and sequencing of cDNAs coding for two distinct pea chloroplast aldolases, Arch. Biochem. Biophys. 298 (1992) 192 – 197.
[19] K. Tsutsumi, Y. Kagaya, S. Hidaka, J. Suzuki, Y. Tokairin, T. Hirai, D.L. Hu, K. Ishikawa, S. Ejiri, Structural analysis of the chloroplastic and cytoplasmic aldolase-encoding genes implicated the occurrence of multiple loci in rice, Gene 141 (1994) 215 – 220.
[20] T. Kubo, Tobacco genetic resources and biotechnology, in: T. Watanabe, E. Pehu (Eds.), Plant Biotechnology and Plant Genetic Resources for Sustainability and Pro-ductivity, R.G. Landes, Austin, 1996, pp. 103 – 113. [21] V. Haake, R. Zrenner, U. Sonnewald, M. Stitt, A
moder-ate decrease of plastid aldolase activity inhibits photo-synthesis, alters the levels of sugars and starch, and inhibits growth of potato plants, Plant J. 14 (1998) 147 – 157.
[22] D. Heineke, A. Kruse, U.-I. Flu¨gge, W.B. Frommer, J.W. Riesmeier, L. Willmitzer, H. Heldt, Effect of anti-sense repression of the chloroplast triose-phosphate translocator on photosynthetic metabolism in transgenic potato plants, Planta 193 (1994) 174 – 180.