A comparison of stilbene and chalcone synthases including a new
stilbene synthase gene from
Vitis riparia
cv. Gloire de Montpellier
P.H. Goodwin
a,*, T. Hsiang
a, L. Erickson
baDepartment of En6ironmental Biology,Uni6ersity of Guelph,Guelph,Ont.,Canada N1G2W1 bDepartment of Plant Agriculture,Uni6ersity of Guelph,Guelph,Ont.,Canada N1G2W1
Received 13 May 1999; received in revised form 30 August 1999; accepted 30 August 1999
Abstract
A stilbene synthase gene was cloned fromVitis ripariacv. Gloire de Montpellier after PCR amplification with primers designed to include the start and stop codons of stilbene synthase genes ofV.6inifera. The exon was very similar to that of other stilbene
synthases, particularly those from V.6inifera (99% nucleotide identity). An intron was found which interrupted the predicted codon for cysteine in the same location as in other stilbene and chalcone synthase genes. The intron showed high nucleotide identity (86%) with an intron from a stilbene synthase gene ofV.6inifera. TheV.ripariasequence was used in an evaluation of the relatedness of stilbene and chalcone synthases of plants. Five procedures involving distance, parsimony and maximum likelihood methods were used for constructing phylogenetic trees, and they yielded slightly to considerably different results. However, none of the trees were consistent with a previous hypothesis that stilbene and chalcone synthases cluster solely based on the genetic relatedness of the species, implying that stilbene synthase genes arose independently in plant families. In our analyses, stilbene and chalcone synthases ofVitisalways clustered separately. The relatedness and origin of stilbene and chalcone synthases appears to be more complex than originally believed. © 2000 Elsevier Science Ireland Ltd. All rights reserved.
Keywords:Stilbene synthase; Chalcone synthase;Vitis riparia
www.elsevier.com/locate/plantsci
1. Introduction
Stilbenes are phenylpropanoid derivatives pro-duced by plants. The key enzyme for their synthe-sis is stilbene synthase, which catalyzes the addition of malonyl-CoA to 4-coumaroyl-CoA or cinnamoyl-CoA [1]. Stilbenes can be produced constitutively in wood of trees but can also be induced by plant stresses, such as wounding, ozone damage or disease [1]. Stilbenes are toxic to fungi and may contribute to disease resistance as phytoalexins. One reason for an increased interest in stilbene synthases in recent years is that the substrates for stilbene synthase are found widely in plants, and transfer of these genes to plants lack-ing stilbene synthase can increase their disease
resistance. Expression of stilbene synthases has been reported in transgenic barley, oilseed rape, potatoes, rice, tobacco, tomatoes and wheat, and it has increased significantly disease resistance to a number of phytopathogenic fungi [2 – 6].
The cloning and sequencing of stilbene synthase genes revealed that they have many similarities to chalcone synthases, which are involved in flavonoid synthesis [7]. This appears to be related to the fact that both enzymes use the same sub-strates and share the same basic mechanism of catabolism [1]. However, each enzyme yields dif-ferent products, either stilbenes or naringenin chalcone, which differ in their ring structures. A comparison of stilbene and chalcone synthases showed 70 – 75% amino acid homology, an intron in a conserved location, and a conserved cysteine which is essential for the activity of both enzymes [1,8]. This has raised questions as to the evolution-* Corresponding author. Tel.: +1-519-824-4120, ext. 2754; fax:
+1-519-8370442.
E-mail address:[email protected] (P.H. Goodwin)
ary origins and relatedness of chalcone and stil-bene synthases. Studies of two different stilstil-bene synthases ofPinus syl6estris showed that they had
higher homology to chalcone synthase of P.
syl6estris, than they did to the stilbene synthases of
other plants [9,10]. Also, a consensus phylogenetic tree of chalcone and stilbene synthase proteins showed that stilbene synthases did not form a separate cluster but rather grouped with the chal-cone synthases of related families [11]. Since that report, there have been additional reported se-quences, including chalcone synthase sequences
from Vitis 6inifera and a new stilbene synthase
from V. riparia reported here, that were used in
our analyses.
Thus far, stilbene synthase genes have been cloned from a very limited number of plants, including two varieties of cultivated grapes (V.
6inifera), peanut (Arachis hypogaea) and two
spe-cies of pines (P. strobus and P. syl6estris)
[7,9,10,12 – 15]. The goal of this research was to re-examine the relatedness of stilbene synthases and chalcone synthases and to include a newly cloned stilbene synthase gene obtained from an-other species of Vitis, V. riparia. In our analyses, several different tree construction procedures in-volving distance, parsimony and maximum likeli-hood methods were used, and the results were compared.
2. Materials and methods
DNA from leaves of V. 6inifera cv. Optima and
V. riparia cv. Gloire de Montpellier was extracted
according to Lodhi et al. [16]. Twenty-five nanograms of extracted DNA was used for PCR. Amplification was done in a total volume of 25 ml using the primers STSYF (5%-TGGG(T/A)
TCAATGG(T/C)TTCAG) and STSYR (5%
-CACTTAATTTGTCACCA(A/T)AGGA). These
primers were designed based on relatively highly conserved nucleotide sequences at the start and stop codons, respectively, of stilbene synthases of
V. 6inifera ‘Optima’ and V. 6inifera ‘Labrusca a
Foglia Frastagliata’, and on highly conserved se-quences of the stilbene synthases ofP. strobusand
P. syl6estris corresponding to the 6 – 8 nucleotides
near the 3% end of the primers. The amplification
reaction was composed of 200mM dNTPs, 0.5mM primers, 0.03 U/ml of Taq DNA polymerase
(Gibco BRL, Gaithersburg, MD), 1.5 mM MgCl2
and 1×Taq DNA polymerase buffer. The cycling parameters were: 94°C for 5 min followed by 30 cycles at 94°C for 45 s, 50°C for 45 s and 72°C for 45 s, and a final extension time at 72°C for 7 min. The PCR fragment was cloned into T-vectors constructed from pBluescriptII KS (Stratagene, LaJolla, CA) according to Hadjeb and Berkowitz [17]. Restriction enzymes and related reagents were purchased from Pharmacia Biotech Inc., and were used according to the manufacturer’s instruc-tions. Plasmid DNA was introduced into com-petent Escherichia coli by transformation [18]. Sequencing was done with an ABI Prism model 377 DNA sequencer (Applied Biosystems Inc., Foster City, CA).
2.1. Sequence comparisons
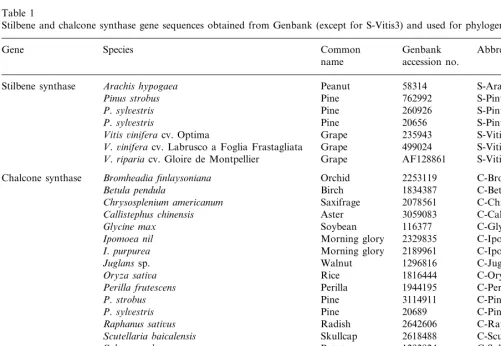
Amino acid sequence comparisons were made between 17 chalcone synthases and seven stilbene synthases including our sequence from V. riparia
(Table 1). The sequences were aligned with
CLUSTAL-W [19] using default parameters.
Includ-ing gaps, a 410-amino acid length was aligned. Phylogenetic trees were then constructed using distance, parsimony and maximum likelihood methods. For all three methods, the aligned se-quences were first subjected to bootstrapping using the program SEQBOOT in the PHYLIP package
[20] (available from http://evolution.genetics. washington.edu/phylip.html). Genetic distances within 1000 bootstrap replicates were calculated with the PHYLIP program PROTDIST using a
Day-hoff PAM matrix. The distance matrices were then analyzed with three different procedures: (1) with the PHYLIP program NEIGHBOR using the
neigh-bor-joining algorithm; (2) with the PHYLIP
pro-gram NEIGHBOR using the UPGMA algorithm;
and (3) with the PHYLIP program FITCH. A
parsi-mony method implemented in thePHYLIPprogram PROTPARS was used also to examine 1000
boot-strap replicates of the aligned sequences. The aligned sequences were also analyzed using 50 bootstrap replicates with a maximum likelihood method in the program PROTML (part of a
com-puter package called MOLPHYwhich was compiled
The multiple data sets from the five procedures of the three methods were then separately analyzed with the PHYLIP program CONSENSE to obtain
bootstrap values reflecting the consistency of tree branching patterns, and dendrograms were pro-duced with the PHYLIP programDRAWGRAM. The
chalcone synthase-like protein sequence from
Streptomyces griseus, a prokaryote, was used to
root all trees.
3. Results
Primers were designed which would amplify the full length of a stilbene synthase gene. Amplifica-tion of genomic DNA fromV. 6inifera cv. Optima
and V. riparia cv. Gloire de Montpellier both
yielded a single band of 1550 bp (data not
shown). Cloning and sequencing of the stilbene synthase gene from V. riparia (Genbank accession no. AF128861) revealed that it had 99% nucleotide
identity to the stilbene synthases of V. 6inifera but
only 63% identity to the chalcone synthases of V.
6inifera (Fig. 1). Nucleotide identity of the V.
riparia stilbene synthase gene was 65, 63 and 62%,
respectively, with the stilbene synthases of A. hy
-pogaea, P. strobus and P. syl6estris. An
examina-tion of the predicted protein sequence of the V.
riparia gene showed that the sequence motif,
GVLFGFGPGLT, which is the family signature sequence for stilbene and chalcone synthases [9], was present (Fig. 1). Furthermore, the sequence motif, IPNSAGAIAGN, which is specific for stil-bene synthases [15], was found also in the pre-dicted protein sequence of theV. ripariagene (Fig. 1). The size of the predicted protein was 42716 daltons with an isoelectric point of 5.81. One intron was present which interrupted the codon for cysteine at the same location as reported in all other stilbene and chalcone synthase genes. The intron sequence was compared with that of two stilbene synthase genes of V. 6inifera cv. Optima
Table 1
Stilbene and chalcone synthase gene sequences obtained from Genbank (except for S-Vitis3) and used for phylogenetic analyses
Species
Gene Common Genbank Abbreviation
accession no.
Vitis6inifera cv. Optima Grape 235943
V.6inifera cv. Labrusco a Foglia Frastagliata Grape 499024 S-Vitis2
V.ripariacv. Gloire de Montpellier Grape AF128861 S-Vitis3
Bromheadia finlaysoniana Orchid
Chalcone synthase 2253119 C-Bromhead
C-Betula
Birch 1834387
Betula pendula
Chrysosplenium americanum Saxifrage 2078561 C-Chrysosplen
Callistephus chinensis Aster 3059083 C-Callistephus C-Glycine 116377
Soybean
Glycine max
Ipomoea nil Morning glory 2329835 C-Ipomoea1
I.purpurea Morning glory 2189961 C-Ipomoea2 C-Juglans Walnut 1296816
Juglanssp.
Oryza sati6a Rice 1816444 C-Oryza
Perilla frutescens Perilla 1944195 C-Perilla
P.strobus Pine 3114911 C-Pinus1
P.syl6estris Pine 20689 C-Pinus2
2642606 Radish
Raphanus sati6us C-Raphanus
Scutellaria baicalensis Skullcap 2618488 C-Scuttellaria
Solanum tuberosum Potato 1292924 C-Solanum Grape
V.6inifera cv. Cabernet Sauvingnon 3288721 C-Vitis1
V.6inifera Grape 2465406 C-Vitis2
3702261 Actinomycete
Streptomyces griseus C-Streptomyces
Fig. 1. Nucleotide sequence alignment of the coding regions of a stilbene synthase ofV.riparia(NS-Vitis3), a stilbene synthase of V. 6inifera (NS-Vitis1) and a chalcone synthase of V. 6inifera (NC-Vitis2). Nucleotides corresponding to the amino acid
signature sequences for stilbene synthases are shaded in reverse, and the stars below each line of alignment indicate fully conserved sites.
(Fig. 2) The homology was very high (86.2% iden-tity) with the similarly-sized intron of the Vst1 gene ofV. 6inifera cv. Optima, but there was only
53.7% identity with the intron of the Vst2 stilbene synthase gene. There were four differences in the 21-bp region of the intron in the V. riparia gene that had been found to be identical between the
Vst1 and Vst2 genes [15].
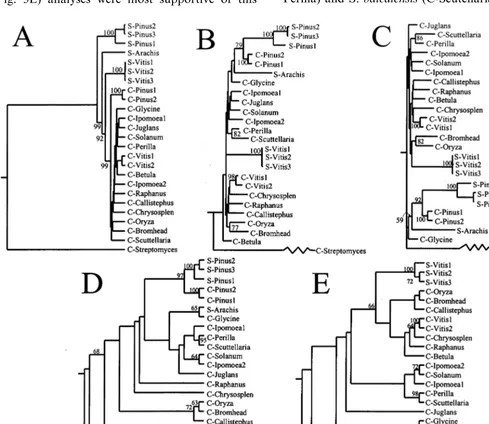
Alignment of the amino acid sequence of the stilbene synthase of V. riparia with that of other
stilbene and chalcone synthases was done with five different procedures (Fig. 3) involving distance, parsimony and maximum likelihood methods. Us-ing a parsimony method, Tropf et al. [11] reported that stilbene synthases did not cluster separately from chalcone synthases but both grouped to-gether and could be subdivided based on the sys-tematics of the plant families. Our analyses with all five procedures also found that stilbene syn-thases did not form a separate cluster from
cone synthases (Fig. 3). However, UPGMA analy-sis placed all the chalcone synthases, including those from Pinus spp., in one cluster, and all the stilbene synthases of different genera as succes-sively more distant from chalcone synthases (Fig. 3A). In the other types of analyses (Fig. 3B – E), stilbene and chalcone synthases ofPinus spp. clus-tered together, but for the Vitis spp., the stilbene and chalcone synthases did not cluster together. Parsimony (Fig. 3D) and Maximum Likelihood (Fig. 3E) analyses were most supportive of this
distinction with bootstrap values above 68% at the branching points of these two groups. The chal-cone synthase of soybean (C-Glycine) and the stilbene synthase from peanut (S-Arachis), both members of the Fabaceae, did not cluster together in any of the analyses.
In some instances branching patterns reflected the taxonomic position of the species sequenced. For instance, except for UPGMA analysis, the chalcone synthase sequences of P. frutescens (C-Perilla) and S. baicalensis (C-Scutellaria) clustered
together (bootstrap values ranged from 82 to 98%) and both are members of the Lamiaceae. How-ever, the chalcone synthases of I. purpurea clus-tered closer to S. tuberosum than to I. nil for all the types of analysis other than UPGMA, even thoughI. purpureaandI. nilare in the same genus in the Convolvulaceae, whereas S. tuberosum is a member of the Solanaceae.
4. Discussion
Using primers from relatively highly conserved regions at the start and stop codons, we were able to clone a full length copy of a stilbene synthase gene from a species of grapes other than V.
6inifera. Only a single band was obtained from V.
riparia, which was the same size as that amplified
from V. 6inifera cv. Optima. It is unlikely,
how-ever, thatV.riparia has only one stilbene synthase gene. Stilbene synthase genes have been found to be part of multigene families in grapes, peanuts and a number of tree species [7,14,15,21]. At least seven stilbene synthase genes were found in V.
6inifera cv. Optima [15]. Three of these were
closely linked, but the coding regions differed only slightly. However, the sequence of the intron for two of these genes was determined and differed considerably in sequence and in length [15]. At least ten stilbene synthase genes were obtained
fromP.syl6estriswhich could be grouped into five
subclasses based on their intron size [22]. Our primers, therefore, probably only amplified one copy of a stilbene synthase gene since only a single band forV. ripariaand V. 6inifera cv. Optima was
obtained from genomic DNA, whereas multiple bands would be expected if other copies were amplified because of variations in intron lengths. At a protein sequence level it is not easy to distinguish between stilbene and chalcone syn-thases. Stilbene and chalcone synthases share many structural features. One distinction is se-quence surrounding Ser-250, which is –
IPN-SAGAIAGN – in stilbene synthases and
– IPDSAGAIAGD – in chalcone synthases [15]. Schroder and Schroder [1] noted that most of the differences between the two enzymes were in charged amino acids, which may be related to the type of final products. Because the cluster analysis of chalcone and stilbene synthases of Tropf et al. [11] showed that stilbene synthases grouped with
chalcone synthases of related plants, it was sug-gested that there was no ancestral stilbene syn-thase for all plants. Instead, stilbene synsyn-thases were proposed to have arisen independently sev-eral times from chalcone synthases. However, our results do not support this conclusion. Although the different types of analysis did not yield identi-cal branching patterns, the chalcone and stilbene synthases did not cluster together consistently based solely on the systematic groupings of the plants. The chalcone synthases from grapes always clustered with the other chalcone synthases and never with the grape stilbene synthases. No chal-cone synthases from grapes were included in the analysis of Tropf et al. [11]. In the UPGMA analysis, all the chalcone synthases were in a dis-tinct cluster, and the chalcone synthase sequences from a wide variety of monocots and dicots were very similar to each other compared to the more diverse stilbene synthases. However, the other four types of analysis did not support such a strong differentiation of chalcone and stilbene synthases, although none of them supported the hypothesis of Tropf et al. [11].
parsimony or maximum likelihood methods [28]. Distance methods have a major advantage of re-quiring less computation than the other methods [29].
One of the major criteria for evaluating the accuracy of phylogenetic trees is congruence [30]. Congruence, or similarities among branching pat-terns derived from different data sets and using different methods of analysis, are a likely result of phylogeny rather than random chance [31]. In our analyses with five procedures involving three methods, we found that different interpretations are possible with the results of the different meth-ods, even among the three distance procedures. One of the reasons for these disparities is the weakness of the branches as shown by low boot-strap values (Fig. 3). The following interpretations are based on the similarities among all tree topologies.
In all five dendrograms, the stilbene synthases of grapes and peanuts never clustered strongly with those from pines, which may be because stilbene synthases can be divided into two groups: the pinosylvan synthases from pines that prefer cin-namoyl-CoA as a substrate and the resveratrol synthases from grapes and peanuts that preferen-tially utilize 4-coumaroyl-CoA as a substrate, like the chalcone synthases. The pinosylvan synthases from the two pine species always clustered to-gether, even though one of these is unique in preferring dihydroCoA to cinnamoyl-CoA as a substrate [9]. The chalcone synthases of pine also always clustered with the pine stilbene synthases except in the UPGMA analysis. Since these genes fromPinus are the only ones from any gymnosperm in these analyses, it would be ex-pected that they could have a relatively high diver-gence from the stilbene and chalcone synthases found in angiosperms.
All stilbene and chalcone synthases, including the one inV. riparia, have a single intron splitting a cysteine codon at a exactly the same position, which further supports the similarity between the genes of the two enzymes [17]. The intron se-quence in V. riparia is more similar to the intron of theVst1 stilbene synthase gene inV.6inifera cv.
Optima than are the Vst1 and Vst2 stilbene syn-thase genes to each other, even though they are from the same genome [15]. The only notable similarity in the Vst1 and Vst2 introns in V.
6inifera cv. Optima was an identical 21 nucleotide
sequence, which is different in the V. riparia se-quence. Because so few stilbene synthase gene have been studied, it is difficult to assess the importance of this similarity in the introns. Per-haps it is related to the fact that stilbene synthases exist in multi-gene families, where the amino acid sequences show very few and mostly conservative changes, but the promoters and introns are highly variable [15]. There may be similar promoters and introns for different stilbene synthases of V.
6inifera and V.riparia, and these may be linked to
the different patterns of expression. In V. 6inifera
cv. Optima, two of the stilbene synthase genes differ by 100-fold in their response to a pathogen-derived elicitor of plant defense responses [15]. The primers used in this experiment may have selected for a particular member of the stilbene synthase gene family with this intron.
The stilbene synthase of V. riparia was very similar to those of the two cultivars of V. 6inifera.
V. 6inifera is a Eurasian species that is widely
planted as a fruit crop, and V. riparia is a North American species found mostly in southern Canada and the mid-western and eastern states bordering Canada. Unlike V. 6inifera, V. ripariais resistant to several diseases of North American origin, such as downy mildew, powdery mildew and black rot, and the cultivar Gloire de Montpel-lier is a selection of V. riparia that is resistant to phylloxera and has been used as a commercial rootstock for over a century [32]. Despite these differences, the genes are extremely similar, and it appears that the stilbene synthase gene has changed little from the ancestral grape species that evolved into these two species.
Acknowledgements
We would like to thank Helen Fisher at the Horticultural Research Institute of Ontario for supplying the plant material and for technical assistance from Jenny Shih and Julianne Gerspacher. This research was supported by the Ontario Soybean Growers Marketing Board and the Ontario Ministry of Agriculture, Food and Rural Affairs.
References
[1] J. Schroder, G. Schroder, Stilbene and chalcone synthases: related enzymes with key functions in plant-specific path-ways, Z. Naturforsch. 45c (1990) 1 – 8.
[2] R. Hain, H.J. Reif, E. Krause, R. Langebartels, H. Kindl, B. Vornam, W. Wiese, E. Schmelzer, P.H. Schreier, R.H. Stocker, K. Stenzel, Disease resistance results from foreign phytoalexin expression in a novel plant, Nature 361 (1993) 153 – 156.
[3] G. Leckband, H. Lorz, Transformation and expression of a stilbene synthase gene ofVitis6iniferaL. in barley and wheat for increased fungal resistance, Theor. Appl. Genet. 96 (1998) 1004 – 1013.
[4] J.E. Thomzik, Transformation in oilseed rape (Brassica napusL.), in: Y.P.S. Bajaj (Ed.), Biotechnology in Agricul-ture and Forestry, vol. 23, Springer-Verlag, Berlin, 1993, pp. 170 – 182.
[5] J.E. Thomzik, Gentransfer in plants, Pflanzenschutz Nachr. Bayer 49 (1996) 5 – 23.
[6] J.E. Thomzik, K. Stenzel, R. Stocker, P.H. Schreier, R. Hain, D.J. Stahl, Synthesis of a grapevine phytoalexin in transgenic tomatoes (Lycopersicon esculentumMill.) condi-tions resistance against Phytophthora infestans, Physiol. Mol. Plant Pathol. 51 (1997) 265 – 278.
[7] G. Schroder, J.W.S. Brown, J. Schroder, Molecular anal-ysis of resveratrol synthase, Eur. J. Biochem. 172 (1988) 161 – 169.
[8] T. Lanz, S. Tropf, F.J. Marner, J. Schroder, G. Schroder, The role of cysteines in polyketide synthases, J. Biol. Chem. 266 (1991) 9971 – 9976.
[9] J. Fliegmann, G. Schroder, S. Schanz, L. Britsch, J. Schroder, Molecular analysis of chalcone and dihy-dropinosylvin synthase from Scots pine (Pinus syl6estris),
and differential regulation of these and related enzyme activities in stressed plants, Plant Mol. Biol. 18 (1992) 489 – 503.
[10] A. Schwekendiek, G. Pfeffer, H. Kindl, Pine stilbene synthase cDNA, a tool for probing environmental stress, FEBS Lett. 301 (1992) 41 – 44.
[11] S. Tropf, T. Lanz, S.A. Rensing, J. Schroder, G. Schroder, Evidence that stilbene synthases have developed from chalcone synthases several times in the course of evolution, J. Mol. Evol. 38 (1994) 610 – 618.
[12] F. Melchior, H. Kindl, Coordinate- and elicitor-dependent expression of stilbene synthase and phenylalanine ammo-nia-lyase genes in Vitis cv. Optima, Arch. Biochem. Biophys. 288 (1991) 552 – 557.
[13] S. Raiber, G. Schroder, J. Schroder, Molecular and
enzymatic characterization of two stilbene synthases from Eastern white pine (Pinus strobus). A single Arg/His difference determines the activity and the pH dependence of the enzymes, FEBS Lett. 361 (1995) 299 – 302. [14] F. Sparvoli, C. Martin, A. Scienza, G. Gavazzi, C. Tonelli,
Cloning and molecular analysis of structural genes involved in flavonoid and stilbene biosynthesis in grape (Vitis6inifera
L.), Plant Mol. Biol. 24 (1994) 743 – 755.
[15] W. Wiese, B. Vornam, E. Krause, H. Kindl, Structural organization and differential expression of three stilbene synthase genes located on a 13-kb grapevine fragment, Plant Mol. Biol. 26 (1994) 667 – 677.
[16] M.A. Lodhi, Y. Guang-Ning, N.F. Weeden, B.I. Reisch, A simple and efficient method for DNA extraction from grapevine cultivars andVitisspecies, Plant Mol. Biol. Rep. 12 (1994) 6 – 13.
[17] N. Hadjeb, G.A. Berkowitz, Preparation of T-overhangs vectors with high PCR product cloning efficiency, BioTech-niques 20 (1996) 20 – 22.
[18] H. Inoue, H. Nojima, H. Okayama, High efficiency transformation ofEscherichia coliwith plasmids, Gene 96 (1990) 23 – 28.
[19] J.D. Thompson, D.G. Higgins, T.J. Gibson,CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice, Nucleic Acids Res. 22 (1994) 4673 – 4680.
[20] J. Felsenstein,PHYLIP-Phylogeny Inference Package (Ver-sion 3.2). Cladistics 5, 1989, pp. 164 – 166.
[21] S.M. Baker, E.E. White, A chalcone synthase/stilbene synthase DNA probe for conifers, Theor. Appl. Genet. 92 (1996) 827 – 831.
[22] R. Preisig-Muller, A. Schwekendieks, A. Brehm, H.J. Reif, H. Kindl, Characterization of a pine multigene family containing elicitor-responsive stilbene synthase gene, Plant Mol. Biol. 39 (1999) 221 – 229.
[23] S. Kumar, K. Tamura, M. Nei, MEGA: molecular evolu-tionary genetics analysis, Version 1.0. The Pennsylvania State University, University Park, PA, 1993.
[24] P.H.A. Sneath, R.R. Sokal, Numerical Taxonomy, Free-man, San Francisco, CA, 1973.
[25] N. Saitou, M. Nei, The neighbor-joining method: a new method for reconstructing phylogenetic trees, Mol. Biol. Evol. 4 (1987) 406 – 425.
[26] W.M. Fitch, E. Margoliash, Construction of phylogenetic trees, Science 155 (1967) 279 – 284.
[27] D.L. Swofford, G.J. Olsen, P.J. Waddell, D.M. Hillis, Phylogenetic inference, in: D.M. Hillis, C. Moritz, B.K. Mable (Eds.), Molecular Systematics, 2nd ed., Sinauer Associates, Sunderland, MA, 1996, pp. 407 – 514. [28] D.M. Hillis, B.K. Mable, C. Moritz, Applications of
molecular systematics: the state of the field and a look to the future, in: D.M. Hillis, C. Moritz, B.K. Mable (Eds.), Molecular Systematics, 2nd ed., Sinauer Associates, Sun-derland, MA, 1996, pp. 515 – 543.
[29] W.H. Li, Molecular Evolution, Sinauer Associates, Sunder-land, MA, 1997.
[30] D.M. Hillis, Approaches for assessing phylogenetic accu-racy, Syst. Biol. 45 (1996) 393 – 414.
[31] F.H. Sheldon, A.H. Bledsoe, Avian molecular systematics, 1970s to 1990s, Ann. Rev. Ecol. Syst. 24 (1993) 243 – 278. [32] R.C. Pearson, A.C. Goheen, Compendium of Grape