Transformation of peanut (
Arachis hypogaea
L.): a non-tissue
culture based approach for generating transgenic plants
V.K. Rohini, K. Sankara Rao *
Department of Biochemistry,Indian Institute of Science,Bangalore560012,India
Received 20 April 1999; received in revised form 30 June 1999; accepted 3 August 1999
Abstract
Transgenic peanut (Arachis hypogaeaL. cv. TMV-2) plants have been produced by a tissue culture-independentAgrobacterium tumefaciens-mediated transformation procedure. Embryo axes of mature seeds with one cotyledon excised were incubated on Murashige and Skoog (MS) gelled medium for 2 days, then pricked randomly with a sterile needle and infected by immersion in a suspension ofAgrobacteriumin Winans’ AB medium that was added with wounded tobacco leaf extract.Agrobacteriumstrain LBA 4404 harboring the binary vector pKIWI105 that carries the genes forb-glucuronidase (GUS) and neomycin phosphotrans-ferase (NPT II) was used for transformation. Following a 16 h infection, 18 h decontamination with cefotaxime and germination and growth on soilrite for 16 days under growth room conditions, the seedlings were transferred to greenhouse. About 3.3% of the seedlings were GUS positive as determined by histochemical assay and by PCR analysis. Molecular characterization of primary transformants as well as the T1and T2generation plants has shown that the method ensured insertion, expression and
inheritance of foreign genes in peanut. © 2000 Published by Elsevier Science Ireland Ltd. All rights reserved.
Keywords: Arachis hypogaea L.; Genetic transformation; Tissue culture-independent method; Agrobacterium tumefaciens; b-Glucuronidase; Neomycin phosphotransferase
www.elsevier.com/locate/plantsci
1. Introduction
The peanut (Arachis hypogaea L.) improvement programs aimed at the development of varieties resistant to diseases such as tikka caused by Cer -cospora arachidicola and Cercosporidium person-atum and rust caused by Puccinia arachidis. Modification of peanut genome using genetic engi-neering methods would facilitate rapid develop-ment of new varieties with traits that confer disease resistance. Majority of legume transforma-tion studies has favored the use of Agrobacterium tumefaciens to generate transgenic plants. How-ever, most strategies utilizing Agrobacterium-medi-ated gene delivery have been performed under in vitro culture conditions and require identification of tissues competent for transformation and
devel-opment of tissue culture systems that efficiently convert these tissues into plants. Recently there has been a great deal of interest in the transforma-tion [1 – 8] and in vitro regeneratransforma-tion [9 – 11,6,12] of peanut because it represents one of world’s most important legume crops and therefore is a target crop for improvement. Nevertheless, generalities for peanut regeneration response patterns are difficult to formulate. The different genotypes and/
or explant materials may be contributory to the divergent results reported on regeneration sponse. Another problem with current peanut re-generation techniques has been the 4 – 5 month period required to obtain plants from callus. Transformation procedures that minimize or elim-inate the tissue culture component would therefore be advantageous under such circumstances. The goal of the work presented here was to develop a method of transformation for peanut that is quick, cultivar- and tissue culture-independent.
* Corresponding author. Fax: +91-80-3341814/3341683.
E-mail address:[email protected] (K.S. Rao)
2. Materials and methods
2.1. Plant material
Seeds of peanut cultivar TMV-2 were soaked overnight and later surface sterilized with 0.1% mercuric chloride for 5 – 7 min, followed by thor-ough rinses with sterile water. Embryos with one of the cotyledons cut off at the site of attachment to the primary axis were incubated on semisolid Murashige and Skoog (MS) [13] basal medium for two days prior to infection.
2.2. Bacterial strains and 6ectors
The Agrobacterium strain LBA 4404 harboring the binary vector pKIWI105 [14] was used for transformation. The plasmid carries genes for b -glucuronidase (uid A) and neomycin phospho-transferase (npt II) driven by CaMV 35S and nopaline synthase promoters, respectively. The uid A gene lacks a functional bacterial ribosome bind-ing site that prevents its expression in the bacteria. The Escherichia coli strain XL-1 Blue harboring the plasmid pUCGUS121 was used for probe preparation. This plasmid has the uid A gene under the control of CaMV 35S promoter and nos terminator.
2.3. Infection with Agrobacterium and reco6ery of
transformants
The Agrobacterium strain LBA 4404/pKIWI105 was grown overnight at 29 – 30°C in LB medium (pH −7.0) containing 50 mg ml−1 kanamycin.
The bacterial cells were later resuspended in Winans’ AB medium (pH −5.2) [15] and grown for 18 h. Wounded tobacco leaf extract was later added to this suspension. The embryo axes were pricked randomly with a sterile sewing needle and dunked in the suspension of Agrobacterium in Winans’ AB medium. The infection was carried out by gentle agitation at 28 – 30°C. The seedlings were blot-dried, washed thoroughly with 500 mg ml−1 of cefotaxime for
18 h and placed on
autoclaved soilrite for germination under aseptic conditions in capped bottles. After 5 – 6 days, the germlings were transferred to soilrite in pots and the seedlings were allowed to grow under growth room conditions for at least 10 days before they were transferred to the greenhouse. The pots were
initially covered with polythene bags to maintain humidity. The growth chamber was maintained at 26 – 28°C under a 14 h photoperiod with fluores-cent light of intensity 35 mmol m−2 s−1. For each
experiment 50 embryos were infected and the ex-periments were repeated thrice.
Various transformation conditions viz., infec-tion time, effect of acetosyringone and addiinfec-tion of wounded tobacco leaf extracts on transformation efficiency were evaluated.
2.4. Expression of marker genes
GUS enzyme activity was assessed in the tissues sampled from different phases of development of putative transformants following the method of Jefferson [16].
Expression of GUS was also ascertained by Western blotting. Total proteins were extracted from about 1g of leaf tissue of two months old transformants and resolved on a 10% polyacry-lamide gel following the protocol of Laemmli, [17]. Immunostaining of the immobilized proteins was performed using the GUS antibody from Clonetech following the GUS gene fusion system user manual (Clonetech). The expression of thenpt II gene in the genome of the transformed peanut plants was checked by the NPT II nd-PAGE assay performed according to Reiss et al., [18].
2.5. Molecular analysis
Plant genomic DNA was isolated following the method of Dellaporta et al. [19]. Plasmid DNA from A. tumefaciens strain LBA 4404/pKIWI105 and E. coli strain XL-1 Blue/pUCGUS121 was prepared following the method of Sambrook et al. [20].
For PCR analysis of the uid A gene in the genome of transformants, a 21 mer primer [21] — 5% CTG TAG AAA CCC GTG 3% and 5% CAT
TAC GCT GCG ATG GAT CCC 3% that am-plifies a 514 bp fragment was employed. The PCR was performed in a total volume of 50 ml contain-ing 200 ng template DNA, 4 ml milliQ water, 5 ml 10X buffer containing 15 mM MgCl2. 150 mM
For Southern analysis [22], DNA (10 mg) was digested with the appropriate endonuclease, elec-trophoresed on a 0.8% agarose gel, and blotted on a nylon membrane (Hybond, Amersham). Dot
blot was performed with uncut DNA (5 mg). The uidA gene probe was prepared from pUCGUS121 from E. coli strain XL-1 Blue, by releasing a 2.1-kbuidA fragment using BamH1 and EcoR1. This fragment was labeled using the random prim-ing kit supplied by Amersham. Hybridization sig-nals were detected following exposure of X-ray film to the membrane for 16 h at −70°C.
3. Results
3.1. Optimization of culture conditions and de6elopment of transformation protocol
The feasibility of the transformation strategy adopted in the study was initially evaluated by the number of peanut embryo axes germinating into normal seedlings following wounding by excision of one cotyledon and by pricking with a needle, infection withAgrobacteriumand decontamination treatment.
Preliminary experiments to develop transforma-tion protocol involved optimizatransforma-tion of infectransforma-tion time and6irgene induction treatments to enhance
the transformation efficiency. Fig. 1 shows the maximum number of embryos expressing GUS when infection was carried out for 16 and 24 h. Only two to four embryos per ten sampled have expressed GUS when infected for two and four hours. GUS expression increased with increased time of infection. It attained a plateau when the infection was for 16 h. Though the blue sectors appeared were more with 24 h infection, a 16 h period was chosen for further transformation ex-periments because of a reduction in the germina-tion rate observed on infecgermina-tion for 24 h or longer. As the infection treatment given was for an ex-tended period, and that any further exposure of explants to bacteria appeared deleterious, a dis-crete co-cultivation step that generally follows in-fection was not included in the protocol.
The results of different 6irgene induction
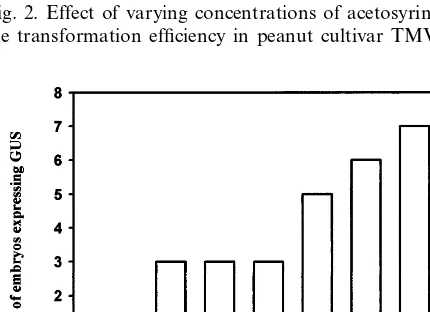
treat-ments are shown in Figs. 2 and 3. Infection of peanut withAgrobacteriumpreviously treated with acetosyringone did not improve transformation efficiency (Fig. 2). Nevertheless, wounded tobacco leaf extract added to the AB induction medium enhanced the transformation efficiency in terms of the number of embryos expressing GUS. Fig. 3 shows the amount of tobacco leaf extract required to obtain optimal infection conditions.
Fig. 1. Effect of infection time on the transient GUS expres-sion in peanut cultivar TMV-2.
Fig. 2. Effect of varying concentrations of acetosyringone on the transformation efficiency in peanut cultivar TMV-2.
Fig. 4. GUS expression in the tissues of peanut cultivar TMV-2 explanted at various stages of recovery of transgenic plants. (a) Embryo axis and the plumule, five days after infection (bar=0.5 mm); (b) embryo axis of peanut cultivar JL-24 (bar=4.5 cm); (c) leaflet from a month old plant (bar=3.5 cm); (d) embryos with one cotyledon removed (T0, primary transformant; T1, progeny)
(bar=4.0 cm).
Fig. 5. Greenhouse established peanut transformants in peanut cultivar TMV-2. (a) A fertile plant of T0generation; (b) plants of
Fig. 6. Expression of neomycin phosphotransferase II gene. (a) Expression in T0plants. Lane 1: total protein extract from
uninfected plant (negative control). Lane 2: total protein extract from a transformant. (b) Expression in T1 plants.
Lane 1: total protein extract from uninfected plant. Lanes 2 and 3: total protein extract from the progeny of transfor-mants.
wounded uninfected as well as infected embryos were germinated in the presence of kanamycin. Therefore, selection on kanamycin was eliminated. Thirty percent of the seeds survived wounding, infection and germinated into healthy plants with 16 h of infection. The seedlings transferred to pots were initially covered with polythene bags while they were grown in the culture incubation room and before they were shifted to the greenhouse. Six primary transformants resulted from three inde-pendent transformation events involving a total of 150 embryos. Five plants, which survived harden-ing and transfer to greenhouse were analyzed for the presence and transmission of the transgenes. These plants have put forth healthy, green and expanded leaves (Fig. 5a). The rate of growth of these infected seedlings was however slow when compared to uninfected seedlings, which might be due to wounding and prolonged infection with Agrobacterium.
3.2. Expression of the npt II gene
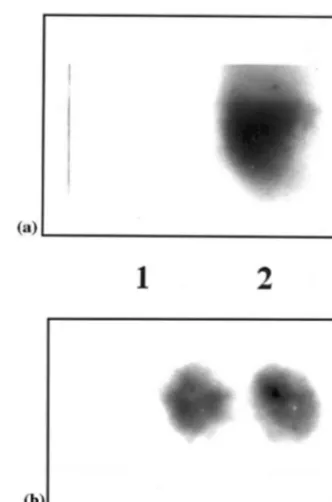
The NPT II assay performed, resulted in a signal at the expected position of 14 kDa with the total protein extracts of the primary transformant and the T1 progeny indicating co-integration,
ex-pression and transmission of npt II gene (Fig. 6a and b). There was no signal seen in case of unin-fected peanut plants.
3.3. Expression and inheritance of the uid A gene
The susceptibility response of embryo axes of peanut to Agrobacterium infection that was deter-mined initially by scoring the transient GUS activ-ity five days after infection, resulted in blue color development throughout the embryonal axes (Fig. 4a). The advantage of eliminating the background GUS activity resulting from bacterial presence in the tissue was conferred by the binary vector pKIWI105. The Agrobacterium LBA 4404 was found to be the most effective strain on peanut cultivars tested. Deep blue sectors were seen occu-pying most part of the leaflet area (Fig. 4c). GUS activity was expressed in various organs and tis-sues of the T0, T1(Fig. 4d), and T2plants. None of
the control plants expressed GUS.
The expression of uid A gene assayed in the greenhouse-established plants by Western blotting has shown a protein band at the expected position
Fig. 7. Western blot analysis ofb– glucuronidase in T0plants.
Lane 1: purified GUS protein (20 mg) (Clonetech) positive control. Lane 2: total protein extract (50mg) from a transfor-mant. Lane 3: total protein extract (50mg) from an uninfected plant.
Winans’ medium (100 ml) added with wounded tobacco leaf extract (2 g in 2 ml sterile water) and a 16 h infection time were used for all the subse-quent transformation experiments. Embryos of peanut cv. TMV-2 when infected in the absence of acetosyringone or tobacco leaf extract did not show GUS expression.
In the experiments performed initially to deter-mine the tolerance to kanamycin, it was observed that uninfected embryos (control) did not germi-nate beyond 150 mg ml−1 of kanamycin. Further,
Fig. 8. PCR of peanut cultivar TMV-2, T0plants using a 21
mer primer, which amplifies a 514-bp uid A gene fragment. Lane 1: pKIWI105 DNA (positive control). Lane 2: DNA from the leaves of uninfected plant. Lanes 3 – 7: DNA from the leaves of T0plants.
of 74 kDa in the total protein profile (Fig. 7). This band was not detected by the antibody in the total proteins of uninfected plants.
3.4. Molecular analysis of the T0 plants
In PCR analysis using primers for theuidA gene, DNA fragments of expected size of 514 bp in length were amplified from the total DNA of the putative transgenic plants (Fig. 8). These DNA fragments were not detected in the DNA of uninfected plants. Dot blot analysis of DNA samples of the five PCR positive plants further confirmed the presence of the uid A gene in the primary transformants (Fig. 9). Southern blot of uncut DNA of one of these plants gave a hybridization signal with a 2.1 kbuid A gene probe. This DNA when digested with Sma I gave a signal at the 10 kb position (Fig. 11, lanes 1 and 7). A hybridization signal was not obtained with the DNA from the non-transformed plant (Fig. 11, lanes 6 and 10). The protocol facilitated recovery of transformants in as short a time as 4 weeks.
3.5. Molecular analysis of the T1 and T2 plants
Seeds of T0 plants (Table 1) were germinated in
the greenhouse (Fig. 5b) and DNA was prepared from leaves of all the plants that germinated. PCR analysis of 52 DNA samples showed the presence of the uidA gene in 36 samples (Fig. 10). These 36 DNA samples gave a hybridization signal in the
Fig. 9. DNA (5mg) from the five PCR positive T0plants was
loaded on a dot blot apparatus and probed with a 2.1-kbuid
A gene fragment. Lanes 1 – 5: DNA from the five PCR positive T0 plants. Lane 6: positive control (pKIWI105
DNA). Lane 7: uninfected plant DNA (negative control).
Fig. 10. PCR to show the inheritance ofuidA transgene in the T1generation. Lane 1: amplified product size of a 514-bpuidA
Fig. 11. Southern blot of T0and T1plants. DNA (10mg) from
the T0 and T1 plants was digested withSma I and probed
with a 2.1-kbuidA gene fragment. Lanes 1 and 7: uncut and
SmaI digested DNA of a primary transformant (T0)
respec-tively. Lanes 2 – 5: uncut DNA of T1plants. Lanes 6 and 10:
uncut and digested DNA of uninfected plant (negative con-trol). Lanes 8, 9, 11 and 12: DNA of T1plants digested with
SmaI.
Fig. 12. Southern blot of T2 generation plants (dot blot).
Uncut genomic DNA (5 mg) was loaded on a dot blot apparatus and probed with a 2.1-kb uid A gene fragment. Lanes 1 – 32: DNA of T2plants.
Abnormal ratio was observed for the T0−1 plant,
which shows its chimeric nature. These data confi-rmed the inheritance and integration of the uid A gene in both T1 and T2 generations.
To demonstrate that the method is genotype-in-dependent, the embryos of a few other Indian peanut cultivars viz., JL-24, ICGS-44, ICGV-86564 were similarly infected with LBA 4404/
pKIWI105 and were allowed to germinate. GUS histochemical assay performed gave intense blue coloration throughout the embryo axes in all these cultivars (Fig. 4b) indicating that the method can be applied to other cultivars which are susceptible to A. tumefaciens infection.
4. Discussion
A potential limitation of current peanut trans-formation is the requirement for effective in vitro regeneration protocols, which are genotype-inde-Dot blot analysis (data not shown) indicating
in-heritance of the uid A gene in the progeny. Southern hybridization of four PCR positive T1
plants was carried out in order to further confirm uidA gene inheritance and integration. Undigested DNA from all the four T1 plants gave a signal at
around 23 kb indicating the integration of the uid A gene. Variation in hybridization pattern was obtained in the case of Sma I digested DNA confirming integration of the gene (Fig. 11). Five T1plants monitored, produced fertile seeds. These
seeds were germinated and assayed for GUS ex-pression (Table 1). Dot blot analysis performed with the DNA of T2plants agreed with the results
of GUS histochemical assay (Fig. 12).
A 3:1 segregation ratio for GUS expression was observed in most of the T1and T2plants (Table 1).
Table 1
Inheritance of theuid A gene in the T1and T2generations
GUS−
GUS+
No. of T1plants tested 3: 1 Segregation ratioa
T0plant
pendent. Minimizing the role of tissue culture in the transformation procedure would therefore be advantageous under such circumstances. The present study provides an A. tumefaciens-based transformation protocol that does not involve tis-sue culture for converting the primary transfor-mants into transgenic plants. Embryo axes with one of the cotyledons excised and further wounded by pricking with a needle were subjected to Agrobacteriuminfection and allowed to germinate and grow into plants ex vitro. These plants ex-pressed the transgenes. Chee et al. [23] in soybean, Feldman and Marks [24] in Arabidopsis have ear-lier demonstrated in planta methods of similar nature.
The assessment of transformation by this method was based largely on the uid A gene expression. Since the uidA gene construct used in these experiments precludes GUS expression in the bacteria, this feature was taken as a direct measure of transformation. The 6ir gene induction
treat-ment viz., Winans’ low phosphate AB medium with and without acetosyringone was initially eval-uated for transformation frequency. Both these treatments, nevertheless, did not result in in-creased transformation efficiency in the cultivar studied. Cheng et al. [5] also observed a lack of significant increase in the uidA gene expression in the peanut cv. Valencia A when treated with ace-tosyringone. Wounded tobacco leaf extracts added to Winans’ AB medium in our experiments, how-ever, increased the uid A gene expression signifi-cantly. Further, the time of infection appeared to affect transformation efficiency. It is understand-able because the quantity of phenolics exuding from the wounded embryo axes would be much less as compared to the amount exuding from seedlings or from a well developed (mature) plant. The infection period was therefore prolonged for 16 h. Increase in the uid A gene expression ob-served with addition of wounded tobacco leaf extract suggests that peanut tissues, particularly embryo axes do not release phenolic compounds or other signal molecules in sufficient quantities so as to elicit sufficient 6ir gene induction or release phenolic compounds which are not specific for transcription of 6ir genes. With prolonged
infec-tion, it is likely that the bacteria remain persistent in the apoplast or in the vascular system of the seedlings in which case they might continue to interact with the plant cells and enhance the
chances of transformation. Although the DNA analysis indicated the presence of transgenes within the tissues of transformants, the contami-nating bacterial DNA in these in planta infected seedlings cannot be ruled out. For this reason, the DNA analysis was carried out with only samples that were sufficiently decontaminated before DNA extraction. NPT II expression assays were also performed with similarly treated samples. The method described presents two main advantages: first, it does not require development of a special protocol for plant regeneration and second, one can easily determine its applicability to other peanut varieties and cultivars by simply observing the phenotype of the infected embryos by perform-ing GUS histochemical assay. Incorporation of methods of this nature into transformation strate-gies will accelerate peanut transformation.
References
[1] J.D. Dong, Y.P. Bi, I.S. Xia, Teratoma induction and nopaline synthase gene transfer in peanut, Acta Genet. Sin. 17 (1990) 13 – 16.
[2] C. Lacorte, E. Mansur, B. Timmermann, Gene transfer into peanut (Arachis hypogaea L.) by Agrobacterium tumefaciens, Plant Cell Rep. 10 (1991) 354 – 357. [3] E. Mansur, C. Lacorte, V.G. De freitas, Regulation of
transformation efficiency of peanut (Arachis hypogaea
L.) explants byAgrobacterium tumefaciens, Plant Sci. 99 (1993) 89 – 93.
[4] M. Cheng, R.L. Jarret, J.W. Demski, Z. Li, Expression and inheritance of foreign genes in transgenic peanut plants generated byAgrobacterium-mediated transforma-tion, Plant Cell Rep. 16 (1997) 541 – 544.
[5] M. Cheng, R.L. Jarret, Z. Li, A. Xing, J.W. Demski, Production of fertile transgenic peanut (Arachis hypogaea
L.) plants using Agrobacterium tumefaciens, Plant Cell Rep. 15 (1996) 653 – 657.
[6] P. Ozias-Akins, J.A. Schnall, W.F. Anderson, C. Singsit, T.E. Clemente, M.J. Adang, A.K. Weissinger, Regenera-tion of transgenic peanut plants from stably transformed embryogenic callus, Plant Sci. 93 (1993) 185 – 194. [7] S. Eapen, L. George, Agrobacterium tumefaciens
-medi-ated gene transfer in peanut (Arachis hypogaeaL.), Plant Cell Rep. 13 (1994) 582 – 586.
[8] M. Cheng, D.C.H. His, G.C. Phillips, Recovery of pri-mary transformants of Valencia-type peanut using
Agrobacterium tumefaciens, Peanut Sci. 22 (1994) 82 – 88. [9] C.M. Baker, H.Y. Wetztein, Influence of auxin-type and concentration on peanut somatic embryogenesis, Plant Cell Tissue Organ Cult. 36 (1994) 361 – 368.
[11] P. Ozias-Akins, W. Anderson, C. Holbrook, Somatic embryogenesis in Arachis hypogaea L.: genotype com-parison, Plant Sci. 83 (1992) 103 – 111.
[12] C.M. Baker, H.Y. Wetzstein, Repetitive somatic embryo-genesis in peanut cotyledon cultures by continual expo-sure to 2,4 D, Plant Cell Tissue Organ Cult. 40 (1995) 249 – 254.
[13] T. Murashige, F. Skoog, A revised medium for rapid growth and bioassays with tobacco tissue cultures, Phys-iol. Plant 15 (1962) 473 – 497.
[14] B.J. Janssen, R.C. Gardner, The use of transient GUS expression to develop Agrobacterium-mediated gene transfer system for kiwi fruit, Plant Cell Rep. 13 (1993) 28 – 31.
[15] S.C. Winans, R.A. Kerstetter, E.W. Nester, Transcrip-tional regulation of the 6ir A and 6ir G genes of
Agrobacterium tumefaciens, J. Bacteriol. 170 (1988) 4047 – 4054.
[16] R.A. Jefferson, Assaying chimeric genes in plants, the GUS gene fusion system, Plant Mol. Biol. Rep. 5 (1987) 387 – 405.
[17] U.K. Laemmli, Cleavage of structural proteins during the assembly of the head of bacteriophage T4, Nature 227 (1970) 680 – 685.
[18] B. Reiss, R. Sprengel, H. Will, H. Schaller, A new sensitive method for qualitative and quantitative assay of neomycin phosphotransferase in crude cell extracts, Gene 30 (1984) 211 – 218.
[19] S.L. Dellaporta, L. Wood, J.B. Hicks, A plant DNA minipreparation: version II, Plant Mol. Biol. Rep. 1 (1983) 19 – 21.
[20] J. Sambrook, E.F. Fritsch, T. Maniatis, Molecular Cloning, A Laboratory Manual, 2nd ed., Cold Spring Harbor Laboratory Press, New York.
[21] B.E. Stummer, S.E. Smith, P. Langridge, Genetic trans-formation of Verticordia grandis (Myrtaceae) using the wild-type Agrobacterium vectors, Plant Sci. 111 (1995) 51 – 62.
[22] E.M. Southern, Detection of specific sequences among DNA fragments by gel electrophoresis, J. Mol. Biol. 98 (1975) 503 – 517.
[23] P.P. Chee, A.K. Fober, L.J. Slightom, Transformation of soybean (Glycine maxL.) by infecting germinating seeds withAgrobacterium tumefaciens, Plant Physiol. 91 (1989) 1212 – 1218.
[24] K.A. Feldman, M.D. Marks, Agrobacterium-mediated transformation of germinating seeds of Arabidopsis thaliana: a non-tissue culture approach, Mol. Gen. Genet. 208 (1987) 1 – 9.