Plant Proteomics: Methods and Protocols, edited by Hervé Thiellement, Michel Zivy, Catherine Damerval, and Valerie Mechin, 2006 354. Differential Display Methods and Protocols, second edition, edited by Peng Liang, Jonathan Meade, and Arthur B.
A Review of Current and Future Molecular
Diagnostic Tests for Use in the Microbiology Laboratory
Introduction
- Genotype-Based Methods
The specificity of the primers used in the amplification reaction determines the accuracy of the test result. An example of the different probe patterns that can be obtained with INNO-LiPA Mycobacteria, v2.0.
Microbial Typing
It has been criticized that RAPD is not reproducible between laboratories due to the use of non-strict PCR conditions (41). Allelic variants are assigned for each housekeeping gene based on sequence difference and the resulting alleles for each of the loci examined determine the sequence type of the isolate.
Promising Applications and Some Concerns
- Cystic Fibrosis Associated Infections
- Diagnosis of Bloodstream Infections
- Determination of Bacterial Load
- Molecular Resistance Testing
Another study comparing conventional and molecular methods for detecting bacteria showed that routine cultures often fail to identify bacterial species present in the lungs of patients with CF (52). Nevertheless, rapid PCR-based assays for resistance testing have been introduced in the laboratory and are excellent additional tools, as has been shown for MRSA (65).
Conclusions
1991) Prevention of chronic Pseudomonas aeruginosa colonization in cystic fibrosis by early treatment. 2004) Early detection of Pseudomonas aeruginosa-comparison of conventional versus direct molecular detection (PCR) from adult patients with cystic fibrosis (CF). Molecular detection of antimicrobial resistance. 2004) Detection of methicillin-resistant Staphylococcus aureus directly from nasal swab specimens by a real-time PCR assay.
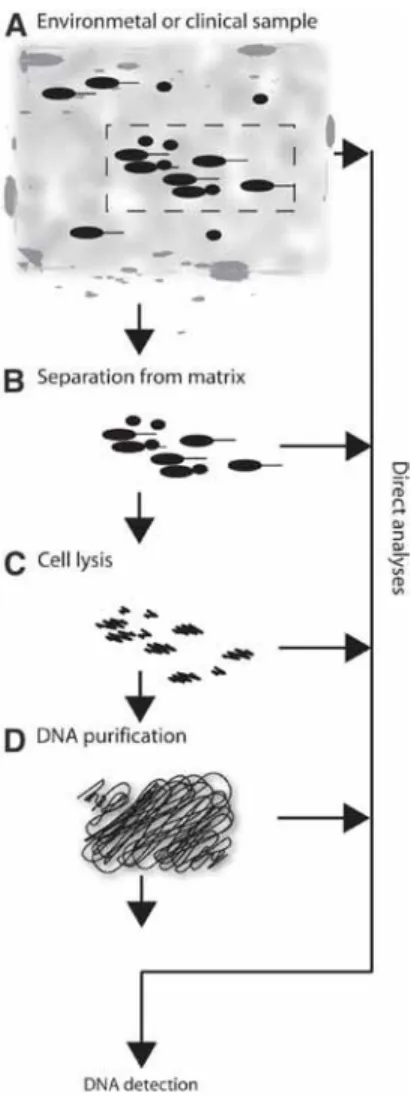
Overview of DNA Purification for Nucleic Acid-Based Diagnostics From Environmental and Clinical Samples
- Sampling
- Separation of Bacteria From Matrix
- Analyses of Crude Lysates
- DNA Purification
- Differentiation Between Viable and Dead Cells
- Future Automation
For soil samples, the separation of the microorganisms from the matrix can be a particular problem. However, standardization of the protocols may be difficult due to the diverse nature of environmental samples.
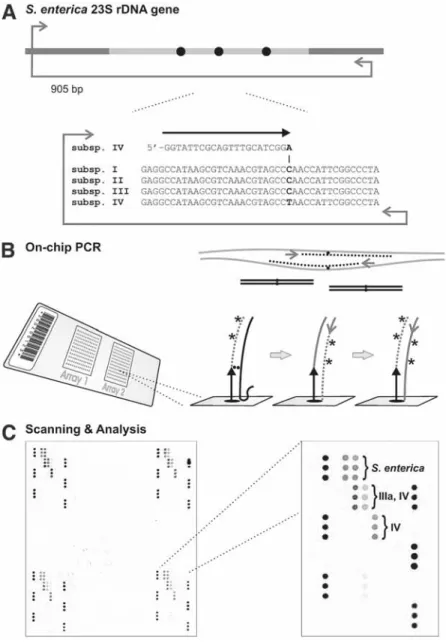
Microarray-Based Detection of Bacteria by On-Chip PCR
Materials
- Primer Design and Synthesis
- Coating of Glass Slides
- On-Chip PCR
- Fluorescence Staining and Scanning
GeneRunner software for melting temperature calculations, sequence manipulation, and solid phase primer design (freely available on the Internet at http://www.generunner.com/). During thermal cycling, a polymerase chain reaction (PCR) product is generated in the liquid phase, which serves as a template (shown as black line) for primer extension in the solid phase.
Methods
- Primer Design and Synthesis
- Coating of Glass Slides 1. Cleaning
- Attachment of Solid-Phase Primers
- On-Chip PCR
- Fluorescence Staining and Scanning 1. Fluorescence Dye Staining
The slides are then thoroughly washed with deionized water and dried with compressed air. Wash slides in 95% ethanol to remove residual silane and dry with compressed air.
Notes
A "high-relicate" array pattern also provides added security in case of possible technical problems, such as scratches on the slide surface that occur during handling of the slides. Whenever possible, stained slides should be stored for at least 2 weeks if the full binding capacity of the slide surface is required.
An Array Biosensor for Detection
- Attachment of NeutrAvidin to the Slide Surface
- Patterning of Biotinylated “Capture” Antibodies
- Assay
- Imaging and Data Analysis
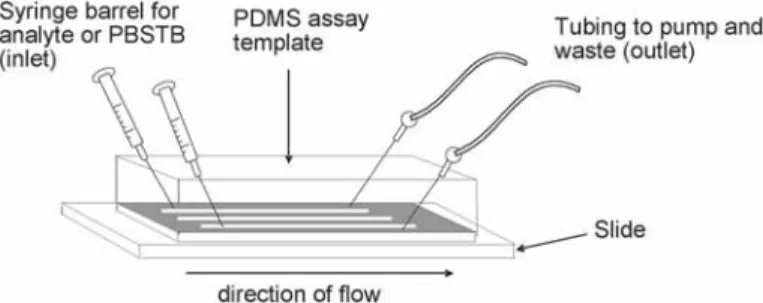
After the third wash, apply a stream of nitrogen to the slide surfaces to remove residual toluene. Place the PDMS patterning template on the NeutrAvid-coated slide surface with channels directed from the slide surface. During optimization, the appropriate concentrations of "capture" and "tracer" antibodies to be used for the assay are determined.
Place the PDMS assay template on the surface of the sample slides with the channels facing the slide and perpendicular to the tracks of the immobilized "capture" antibody. Samples were spiked with SEB, Campylobacter (Campy) or Salmonella (Sal), indicated on the right side of the figure.
Detection of Neisseria meningitidis,
Streptococcus pneumoniae, and Haemophilus influenzae in Blood and Cerebrospinal Fluid
Using Fluorescence-Based PCR
Sample Requirements
For positive and negative controls, culture bacterial cells under optimal conditions for detection of the organism, either on solid or in liquid media. Appropriate media must be used for these organisms when grown in liquid media such as brain-heart infusion broth.
Extraction of Genomic DNA
Fluorescence-Based PCR
1993) Kinetic PCR analysis: real-time monitoring of DNA amplification reactions. 2000) Evaluation of the automated Taqman polymerase chain reaction system for meningococcal DNA detection Applied Bio-systems. 2003) Detection and genotyping of meningococci using a nested PCR approach. 2004) Automation of a fluorescence-based multiplex PCR for laboratory confirmation of common bacterial pathogens. 2005) Interlaboratory comparison of PCR-based identification and genotyping of Neisseria meningitidis. 2003) Comparison of commercial DNA extraction kits for the extraction of bacterial genomic DNA from whole blood samples.
Simultaneous detection of Neisseria meningitidis, Haemophilus influenzae and Streptococcus pneumoniae in suspected cases of meningitis and septicemia using real-time PCR. Use of Hybridization Probes in a Real-Time PCR Assay on the LightCycler® for detection.
Use of Hybridization Probes in a Real-Time PCR Assay on the LightCycler ® for the Detection
- General Equipment 1. Turbidimeter (Densicheck)
- Reagents
- Bacterial Isolation and Preparation of Suspension
- Preparation of Lysozyme Solution
- Sample Preparation
- Real-Time PCR 1. Primers and Probes
- Data Analysis
- Suggested Controls 1. Positive Controls
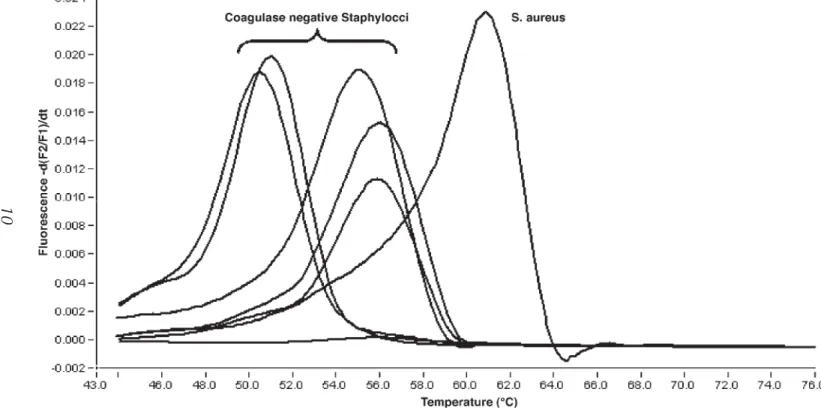
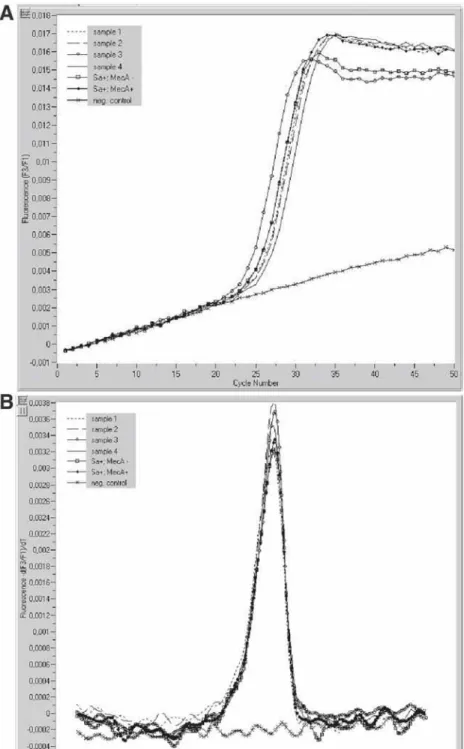
Molecular methods for the rapid identification of MRSA are based on the detection of the mecA and a S. DNA extraction: When using the MagNA Pure LC DNA Isolation Kit III, transfer 200 µL of the lysate to a well of the sample cartridge over and start the automated DNA extraction protocol on the MagNA Pure LC instrument (standard protocol; . see Note 1). After completion of the automated DNA extraction procedure, the eluted DNA (elution volume 100 µL) is ready for the post-elution protocol (see Notes 3 and 4).
After completion of the automated DNA extraction procedure, the eluted DNA (elution volume 100 µL) is ready for the manual preparation of the PCR mixtures (see notes 3 and 4). The following PCR protocol is used for amplification and hybridization based detection of the mecA gene and the S.
Detection of Verotoxin Genes VT 1 and VT 2 in Escherichia coli O157:H7 in Minced Beef
- Enrichment of Minced Beef 1. Modified tryptone soya broth (mTSB)
- Extraction of DNA
- Real-Time PCR
- Enrichment of Minced Beef
- Extraction of DNA
- Real-Time PCR
Place the microcentrifuge tube in the magnetic stand and rotate it by hand until the beads are visibly pulled to the side of the tube. Place the tube in the magnetic stand and rotate it until the beads pull against the side of the tube. Remove 400 µL of the resulting supernatant and add 40 µL of 3 M sodium acetate and 800 µL of absolute ethanol in a 1.5-mL microcentrifuge tube.
Make sure no fat is included when removing a portion of enriched mince, as this will inhibit the binding of the IMS beads. Wear gloves when handling the capillaries and avoid touching the surface of the capillaries as this may affect the fluorescent readings taken by the LightCycler.
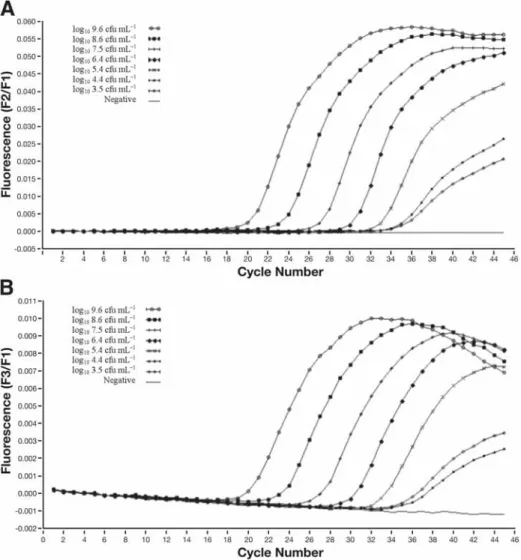
Application of Two-Step Quantitative
Reverse-Transcription PCR to Bacterial Diagnostics
- Preparation of cRNA for Standard Curves 1. Conventional thermocycler
- Preparation of Unknowns 1. Cell density meter
- Data Analysis
- Preparation of cRNA for Standard Curves
- Preparation of Unknowns
- Data Analysis
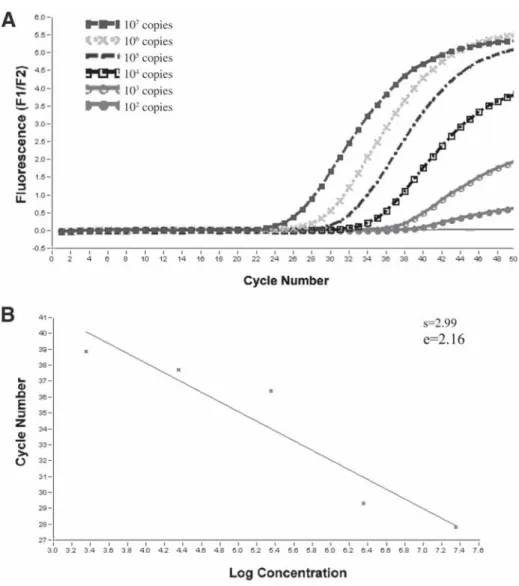
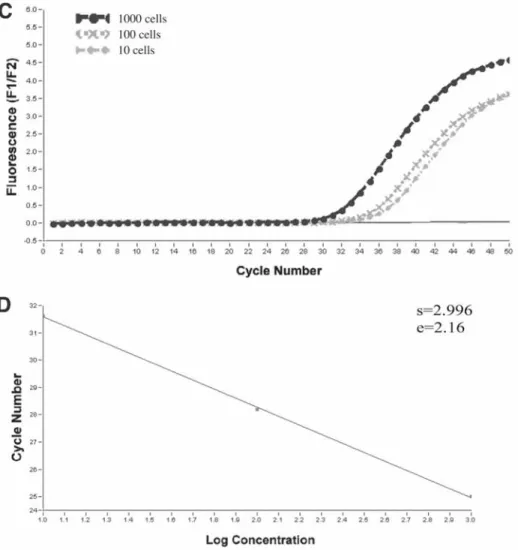
For example, if the amplification efficiency of the standard curve is high, while there is an amplification inhibition. Therefore, in the case of an RNA copy number experiment, it is useful to use RNA in the preparation of the standard curve. Before proceeding with in vitro transcription, it is necessary to identify the orientation of the insert in the plasmid vector.
In vitro transcription is used to generate RNA transcripts consisting of the cloned fragment and a short plasmid sequence. Serial dilutions of the in vitro transcribed RNA are used as template for RT, and this cDNA is amplified using the Light-Cycler.
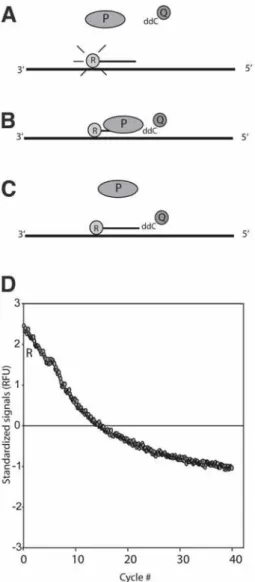
Quencher Extension for Single Nucleotide
Polymorphism Quantification in Bacterial Typing and Microbial Community Analyses
- Template
- QEXT Probes
- Pretreatment of PCR Products
- QEXT Reaction
- Data Analysis
- Pretreatment of PCR Products
- QEXT Reaction
- Data Analysis
- Data Interpretation
The signal is recorded by quenching (reduced fluorescence) of the reporter dye (see Figure 1). The problem with this bacterium is that it is very abundant in the environment and most of the L. The curve decreases proportionally with the uptake rate of the dideoxycytosine per cycle.
The recording rate per cycle is one minus the 'effect'. The uptake rate should be proportional to the amount of template in the sample. The initial development of the QEXT system was done on an ABI PRISM 7700 (Applied Biosystems).
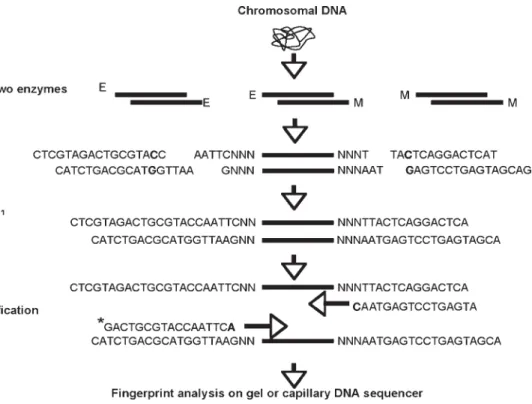
Amplified Fragment-Length Polymorphism and Protein Profiling for Identification
- Isolation of Campylobacter lari
- Isolation of DNA
- Restriction of DNA and Ligation of Adapters
- Acrylamide Gel Electrophoresis for ABI 373A
- Protein Gel Analysis
- Data Analysis
- Bacterial Cultures
- AFLP Analysis 1. Isolation of DNA
- Protein Profile Analysis
Polyacrylamide gel electrophoresis (PAGE) in the presence of the detergent sodium dodecyl sulfate (SDS) has proven to be an important method for analyzing the protein composition of tissues, cells or biological membranes. Analysis of protein profiles, with numerical analysis of limited parts of the profiles leads to clustering of C. cell protein profiles of two representative strains of a C. Roman numbers correspond to clusters defined by numerical analysis of the region indicated by arrows.
Take an image (high resolution) of the gel with a charge-coupled device camera from the lab. Gels are normalized using standard size bands on both sides of the gel.
Use of Peptide Nucleic Acid Probes
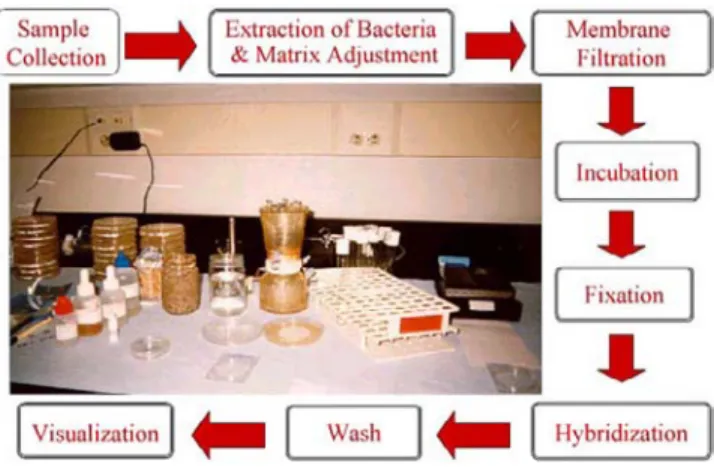
- Sample Collection
- Sample Preparation 1. Extraction of Bacteria
- In Situ Hybridization With PNA Probes
- Detection of Probes
- Quality Assurance
- Collection of Samples
- Preparation of Samples 1. Extraction of Bacteria
- In Situ Hybridization With PNA Probes 1. Fixation of the Microorganisms
- Detection of Labeled Probes
- Quality Assurance
At this point, the microcolonies, barely visible to the naked eye, are formed by a mixed flora on the membrane. For example, if the original probe stock was prepared at a concentration of 2.5 µM, add 3 µl of the stock to 1.5 ml of Hybridization Buffer. Carefully transfer the membrane containing the fixed microbes from the fixation plates to the Petri slides with the probe bacteria facing up.
Immediately submerge the Petri dishes with the membrane in the wash rack containing the wash solution. Using pliers, remove the membrane filter from the wash stand and place it face (the bacteria side) down.
HOOF Prints
Laboratory Requirements
Sample Preparation
PCR Amplification
Processing of Amplicons
However, the custom synthesis of a primer containing NED will be shipped with a report of the total pmol synthesized, from which to calculate the resuspension volume to make a stock solution of 200 pmol/µL.
Sizing of Amplicons by Capillary Electrophoresis
Methods 1. PCR Setup
- Sample Preparation
- PCR Amplification
- Processing of Amplicons: Monitoring Amplification Success
- Sizing of Amplicons by Capillary Electrophoresis With Fluorescent Detection
- Interpretation of Results
- Troubleshooting Potential Problems With the Experimental Data There are a number of things that can affect data production or data interpre-
Be sure that the sample is deposited in the solution at the bottom of the well. The files to load are displayed in the second window at the bottom of the screen. The heights of the peaks (y-axis) are measured in relative fluorescent units (RFUs) emitted as the DNA fragments passed the detector.
The layout of the table is based on the layout of the corresponding polymerase chain reaction plate. To prevent DNA degradation, samples should be stored at 4°C with a pH of 8.0 to 8.5.
Detection of Legionella in Various Sample Types Using Whole-Cell Fluorescent In Situ Hybridization
- Laboratory Equipment
- Reagents and Buffers
- In Situ Detection and Visualization of the Spatial and Temporal Arrangement of Legionella in Protozoa
- Detection and Quantification of Legionella in Water Samples 1. A filtration system (Millipore)
- In Situ Detection and Visualization of the Spatial and Temporal Arrangement of Legionella in Protozoa
- In Situ Detection and Visualization of the Spatial and Temporal Arrangement of Legionella in Biofilms
- Detection and Quantification of Legionella in Water Samples 1. Sterilize the filtration surface of each unit of the filtration system with 100%
In Situ Detection and Visualization of the Spatialand Temporal Arrangement of Legionella in Protozoa and Temporal Arrangement of Legionella in Protozoa. Slides can be analyzed more than once by gently removing the coverslip, rinsing off most of the Citifluor with double-distilled water, air-drying, and storing the slides at –20°C. In Situ Detection and Visualization of the Spatialand Temporal Arrangement of Legionella in Biofilms and Temporal Arrangement of Legionella in Biofilms.
Perform the hybridization as described for in situ detection of Legionella in protozoa (see Subheading 3.1.), adjusting the amount of probe solution of the slide surface. It is recommended that the working solutions be divided into small portions to prevent frequent thawing and freezing of the oligonucleotide probes.
Identification of Diagnostic Proteins
- Computer and Software
- PCR Analysis of Mycobacteria
- Protein Expression–Purification
- Immunoblot Assay 1. Nitrocellulose filters
- Bioinformatic Analysis of the M. paratuberculosis Genome to Identify Candidate Diagnostic Sequences
- Verification of Potentially Diagnostic Coding Sequences
- Production of Heterologously Expressed Proteins From Diagnostic Coding Sequences
Immunological tests for the diagnosis of Johne's disease, such as enzyme-linked immunosorbent assay and gamma interferon production, have historically used complex protein mixtures, including a sonicated whole-cell protein preparation, cell wall preparation, or a purified secreted protein fraction. To address the current lack of sensitivity and specificity in the diagnosis of Johne's disease, we used a comparative genomic approach to identify all M. The development of specific immunoassays for the diagnosis of Johne's disease requires antigens unique to M.
Rapid development of mycobacterial genomics following completion of eight genome sequences (M. paratuberculosis, M. leprae, M. tuberculosis H37Rv and CDC1551, M. bovis AF2122/97 and BCG-Pasteur, M. avium and M. smegmatis [3] –5]) will provide the basis for powerful new identification approaches for a range of specific sequences. By identifying common sequences as well as unique regions within each mycobacterial genome, a better understanding of the genetic requirements required for mycobacterial disease causation will also emerge.