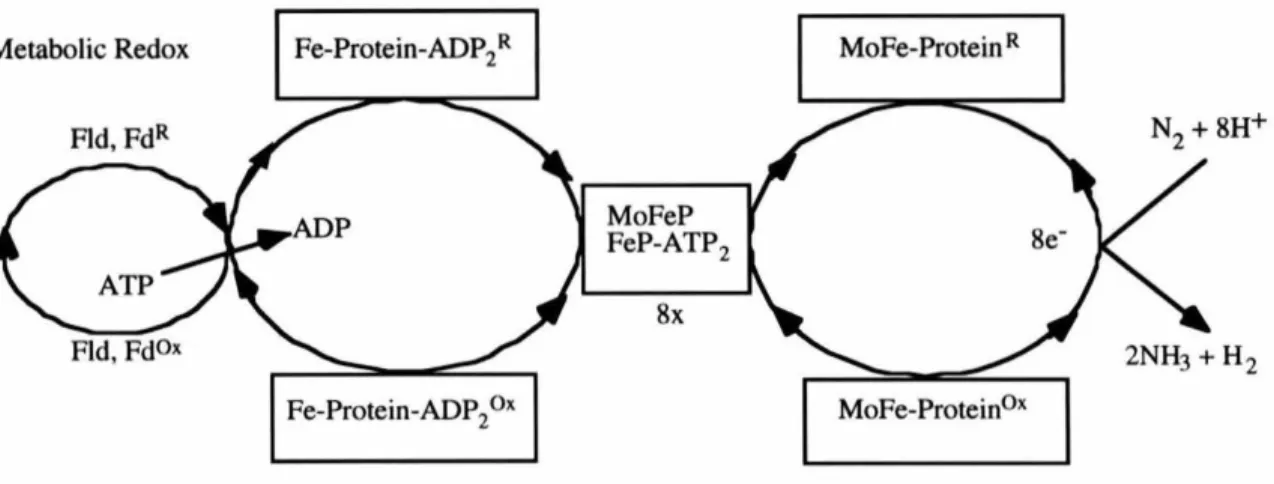
In addition, I am grateful to James Howard, from the University of Minnesota, for excellent and rigorous discussions on nitrogenase. I would also like to thank the past and present members of the Rees lab for their friendship and help. This oxygen-sensitive enzyme, consisting of the separately purifiable nitrogenase iron protein and molybdenum iron protein, couples nucleotide hydrolysis to electron transfer to catalyze the ATP-dependent reaction.
Biological Nitrogen Fixation
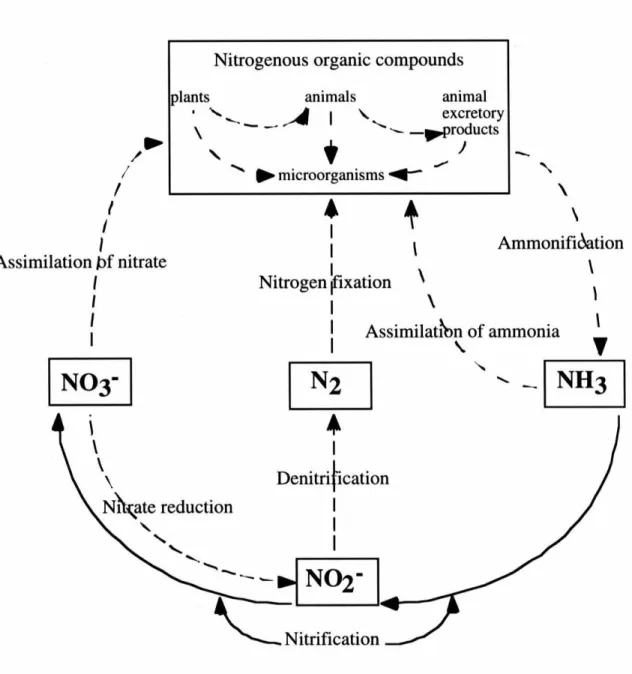
The Nitrogen Cycle
This circulation of solid nitrogen minimizes the dangerous pollutant effects of ammonia entering water sources. Biological nitrogen fixation by nitrogenase provides approximately 60% of the world's newly fixed nitrogen, and will play a critical role in achieving environmentally sustainable agricultural systems. Increasing use of biological nitrogen fixation will reduce the need for nitrogen from commercial fertilizers, with additional benefits in terms of impacts on the global nitrogen cycle, global warming and contamination of ground and surface water (Hardy, 1994).
The Haber-Bosch Process
While the Haber-Bosch process allows large-scale economic production of ammonia for use in fertilizers, explosives and other chemicals (-80 x 109 kg of ammonia per year (Schlesinger, 1991)), biological nitrogen fixation yields even higher annual yields (-170 kg of ammonia per year (Schlesinger, 1991)). x 109 kg ammonia (Schlesinger, 1991)). The industrial production of these fertilizers according to the Haber-Bosch process requires large quantities of natural gas, which releases C02. Food production can therefore indirectly contribute to global warming. Recently, the increased use of biological nitrogen fixation, rather than industrially fixed nitrogen, has been independently identified as a top priority by USAID and UNESCO, with the aim of minimizing disruptions to the natural nitrogen cycle (Hardy, 1994).
Nitrogenase Complex
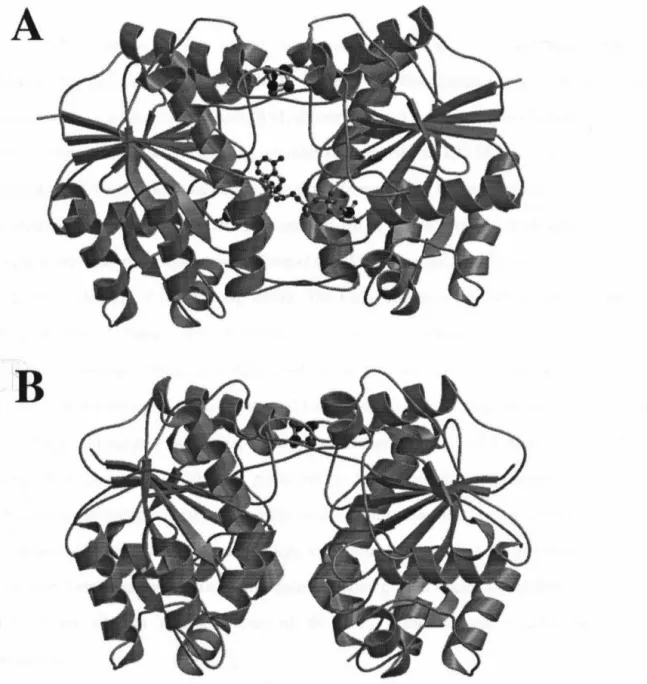
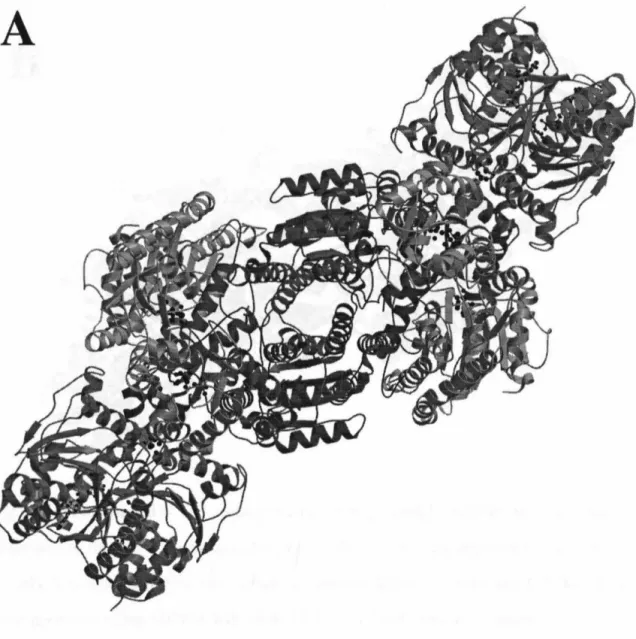
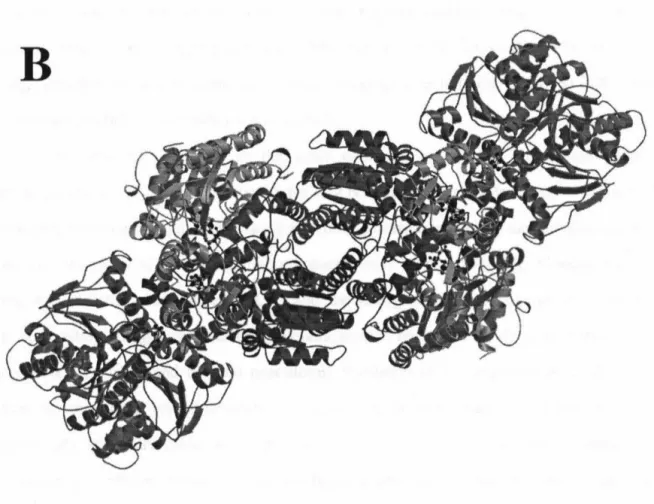
The dihedral axis connecting the two monomers lies vertically in the plane of the page. 34;parallel" bonding mode was recently confirmed in the 3.0 A resolution structure of the nitrogen-nitrogen-stabilized AlF4 complex. The two-fold non-crystallographic axis lies vertically in the plane of the page in (A), perpendicular to the plane of the page in (B), and horizontally in the plane of the page in (C ).
Ball-and-stick model of the lattice packing interaction between molecules in the P212121 Av2 cell. Structure and environment of the metal clusters in the nitrogenase-molybdenum-iron protein from Clostridium pasteurianum. Lanzilotta, WN, Seefeldt, LC (1996) Electron transfer from the nitrogenase iron protein to the [8Fe-(7/8)S] clusters of the molybdenum iron protein.
Elucidation of the mechanism of nucleotide-dependent changes in the redox potential of the [4Fe-4s] cluster in.
Structural Knowledge of Nitrogenase
Research Objective
However, the 2.9 Å resolution of the previous model was insufficient for detailed mechanistic analysis of the structure that could explain the myriad functions of the Fe protein. Comparisons of the A v2 structure with those of Cp2 and the nitrogenase complexes are essential for understanding nitrogenase mechanistic details, such as nucleotide binding, hydrolysis, and complexation, and require near-atomic resolution of uncomplexed Av2. Analysis of this structure and comparisons with other nitrogenase structures places the wealth of available biochemical, genetic and spectroscopic information about nitrogenase in a structural context to improve our understanding of the structural basis of Fe protein function.
Preparation of A v2
Media Requirements
Stock solutions A, B, and C were pipetted into double distilled water in appropriate proportions to create unbuffered media; solid sucrose was added to the mixture. When the solutions had cooled to -30°C, an appropriate portion of stock D was added to the medium; this prevented the formation of phosphate precipitates during autoclaving. Initially, ~1 L of cell culture was inoculated into a 10 L seed fermenter; the cells were then transferred to the 300 L fermenter for final growth.
Purification of Av2
Trizma base (tris(hydroxylmethyl)aminomethane), DNase I (Type IV), and RNase I were from Sigma Chemical (Sigma Chemical, St. Louis, MO). Concentrated dithionite solution was prepared by degassing 5 mL of 0.5 M Trizma base and 4.84 g of N-S O in separate sealed serum bottles (Wheaton, Millville, NJ). A disposable syringe and needle were used to transfer the Trizma to the dithionite-containing vial.
Initial Purification Methods
Luer hubs for disposable needles and 1 cc syringe tips provided the connections between the vacuum/Ar manifold lines and the rubber stoppers on the bottles and flasks. Ultrapure (>99%) Na 2 S 2 O 4 from Noah Technologies (San Antonio, TX) was used as received without further purification.
Pre-Lysis Preparations
Cell Lysis
The bands representing the MoFe and Fe proteins were often overlapping, as were the bands for the Fe protein and the flavodoxin that followed it. The Fe protein fraction was diluted three times without salt buffer and loaded onto column 2. Despite these precautions, complete separation of the yellow flavodoxin from the Fe protein rarely occurred.
Protein Storage and Evaluation
Use of the cannula and sucrose ensured a tight protein band to minimize solution loss from band spreading as the proteins migrated through the column. The Fe protein fraction was concentrated on column 4, a 1 x 10 µm Sepharose CL-6B column, by repeating the steps for column 2.
Final Purification Methods
With the exception of the Superdex 200 column, all columns were freshly packed for each preparation. The silicone tube of the pump was fitted with a stainless steel cannula to pierce the rubber septum of the protein flask. Av2 and Avl were easily separated with the initial gradient; the superior resolution of the Superdex 200 column completely removed flavodoxin from A v2.
Protein Evaluation
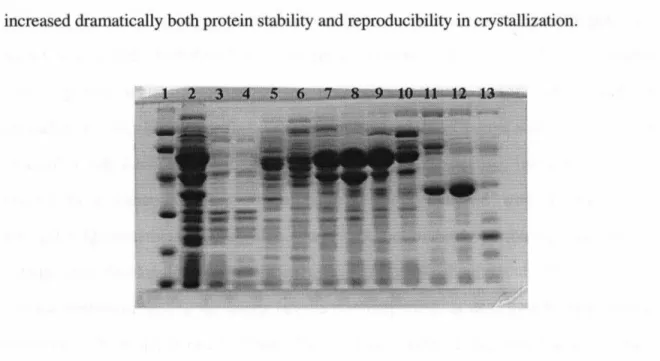
Finally, inclusion of 20% glycerol in the final Av2 buffer dramatically increased both protein stability and reproducibility in crystallization. Crude extract as applied to the column is shown in lane 2, and gel markers (Pharmacia, LMW Marker Kit) are shown in lane 1. SDS PAGE was performed on fractions from each column to monitor protein purity; visual inspection of the final gels indicated that the A v2 used for crystallization was highly purified.
A v2 Crystallization
In the initial crystallization trials, the serum vials were sealed with septa with rubber sleeves and secured around the neck of each vial with 22-gauge copper wire to prevent the septa from coming out during vacuum transfer to the anaerobic chamber. Sealed vials of serum were attached to a degassing manifold using disposable 20-gauge needles inserted through the rubber septum of each vial. Because of the different viscosities of the solutions, the volume losses were expected to differ in each bottle and the crystallization solutions were no longer accurate absolutely or relative to each other.
To circumvent the volume losses inherent in degassing small volumes, a new strategy for preparing Av2 crystallization solutions was devised. Degassed solutions were transferred to the anaerobic chamber via a manual interlock by pumping the interlock down three times to a vacuum of -27 mm Hg and refilling with a 10% H2I balancer gas mixture (Matheson Gas Products). This gas mixture also provided the environment for the anaerobic chamber (residual O2 was removed from the chamber atmosphere by palladium catalysts, which were regenerated 1-2 times per week by heating to -160 ºC outside the chamber for 90 minutes).
Inside the anaerobic chamber, the crystallization solutions were introduced into serum bottles from the stock solutions and sealed as before. Using solutions that were pipetted into the anaerobic chamber resulted in more reproducible crystallizations. Additionally, the availability of large volumes of degassed stock solutions in the anaerobic chamber ensured consistency of solution composition between protein preparations.
Crystallization solutions containing N~Mo0 4 have been found to be unstable over a period of weeks, requiring fresh solutions to be prepared after each protein purification.
Initial Conditions
However, the strong anomalous Fourier signal confirmed the correctness of the Fe4S4 cluster position in the molecular replacement solution. Packing interactions for the P212121 A v2 cell are manifested in the arrangement of several loop regions of the model. Regions of the protein involved in nucleotide hydrolysis (P-loop, Effector I and IT regions) are shown in white.
A loop electron density map for the current structure is shown in Figure 5-7. Not surprisingly, most of the core residues are located in the nucleotide and cluster binding regions. This Fe hybrid protein implicated residues in the deletion loop of Cp2 in the formation of the inactive Cp2-Avl complex.
Role of the nifQ gene product in molybdenum incorporation into nitrogenase in Klebsiella pneumoniae.

Cryocrystallographic Data Collection
Determination of the A v2 Crystal Structure
Molecular Replacement using X-PLOR
Self-Rotation Function
Translation Function
Molecular Replacement using AMORE
NCS Averaging and Solvent Flattening
Improved Phasing Techniques
Current Av2 mode1
Similarity to 2.9 A Resolution Av2
MoPe Protein Binding Surface
Intersubunit Interactions
Di scu ss ion
Crystallization and X-ray Analysis of A v2
Also, the capillary tests were relatively easy and quick to set up in the anaerobic chamber. This was important to counterbalance the drying atmosphere of the anaerobic chamber and the non-trivial challenge of assembling the crystals in the anaerobic chamber. A molybdate anion in each monomer, introduced from the crystallization solution, corresponded to the ~-phosphate position of the nucleotide in the AlF4- complex structure.
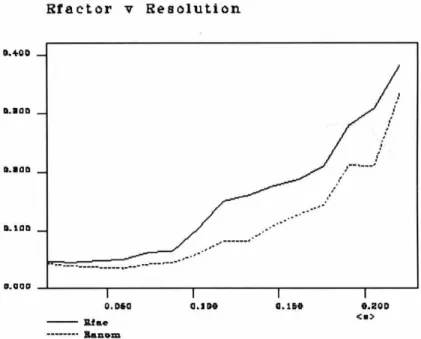
The resulting 0.25-0.75 µm length of melting point capillary was suspended in the wide mouth of the X-ray capillary. Rmerge for the dataset was 0.079 at 2.2 A resolution using SCALEPACK and 0.267 in the highest resolution shell. Analysis of electron density maps did not show the presence of nucleotide in the model.
The search model is rotated and translated in position in the unit cell of the unknown structure until correlation is maximized. The Patterson map to be rotated in the search, Pl, is calculated from the observed intensities. Inspection of the average maps revealed reasonable electron density for many protein side chains, and increased confidence in the original molecular replacement solution.
No interpretable electron density was observed for the three C-terminal residues of subunit A (A287-A289), and these residues are not included in the model. Despite the incorporation of N~MoO4 into the crystallization conditions, examination of electron density maps identified no molybdate ions bound in the P-loop region of the protein, as described for the A v2 structure at 2.9 A resolution. The total temperature factor for non- hydrogen atoms (including solvent molecules) is 31.72 A 2. The average temperature factors for the Fe4S4 cluster and solvent molecules are 20.11 A2 and 46.67 A 2.
Analysis of the A v2 Crystal Structure
Properties of the MgATP and MgADP binding sites on the Fe protein of nitrogenase from Azotobacter vinelandii. Nitrogenase: Properties of the catalytically inactive complex between Azotobacter vinelandii MoFe protein and the Clostridium pasteurianum Fe protein. Repression and derepression of the iron-molybdenum and iron proteins of nitrogenase in Azotobacter vinelandii.