The performance of the primers was assessed for the number of loci they amplified and the number of polymorphic and monomorphic loci. It was found that the GC content of the primers did not affect the primers used in this investigation in any particular way.
INTRODUCTION
INTRODUCTION
Planted forests began to some extent in the 19th century in Europe, and the mid-20th century in North America (Sedjo, 1999). The great variation in the South African environment had a significant influence on the development of forestry in the country.

BLACK WATTLE
- Improvement of black wattle
Both pests cause defoliation of black spruce trees, but can be controlled with insecticides (Govender, 2002). In black weed improvement programs this involves including improved seed in progeny trials.
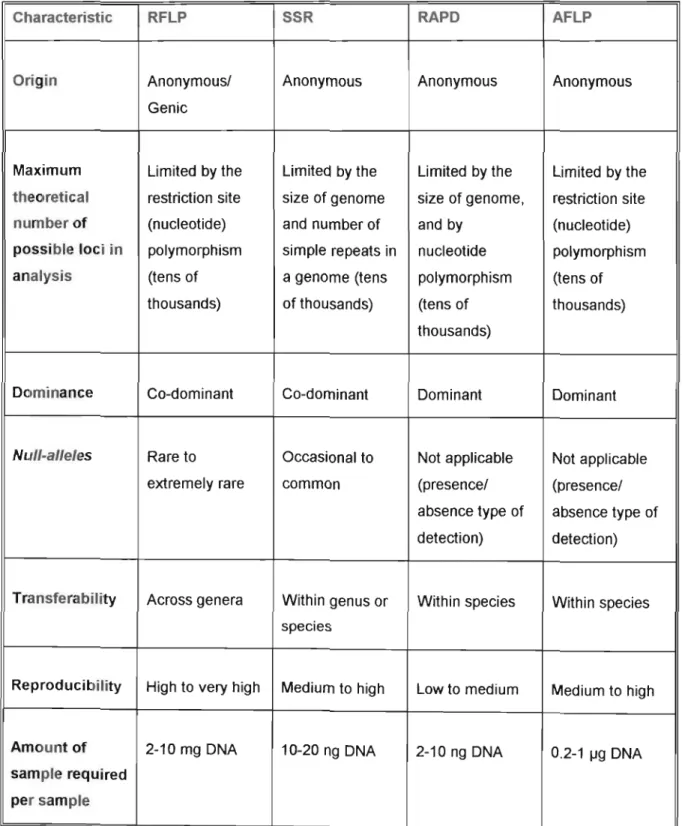
MOLECULAR FINGERPRINTING IN FOREST TREE BREEDING
- Genetic analysis of fingerprints
An allele is represented by a certain number of nuclear sequence repeats (Parker et al., 1998). This initial amplification reduces the total number of confined fragments present in the reaction and is therefore preselective (Questiau et al., 1999).
APPLICATIONS OF FINGERPRINTING IN FOREST TREE BREEDING
In 1995, Bucci and Menozzi investigated the genetic variation of RAPD markers in an Italian population of Norway spruce. Studies include the investigation of genetic variation in natural populations of Acacia mearnsii (Searle and Bell, 1998).
AIMS
Acacia mearnsii was found to exhibit moderate genetic variation (0.201), with the majority (89.2%) of the variation occurring within populations. The level of genetic variation varied considerably between populations, ranging from 0.01 on Ceram Island in Indonesia to 0.21 in Muting, New Guinea.
MATERIALS AND METHODS
INTRODUCTION
MATERIAL
The cells of many plant species contain phenols and polysaccharides that interfere with the process of DNA isolation. Therefore, to optimize the DNA isolation protocol, plant material was collected from trees of different ages (Table 2.2).

METHODS .1 DNA isolation
Add 400 IJI 1 x TE to the DNA pellet in the tube and incubate in a 50 °C water bath for 15 min to dissolve the pellet. Prior to DNA amplification for RAPD fingerprinting, the DNA was evaluated for purity and concentration.

SCORING OF RAPD FINGERPRINTS
In this investigation, each RAPD reaction was repeated twice for each of the identified polymorphic primers and assessed for reproducibility. To analyze the molecular fingerprints, it was necessary to convert the gel fingerprints into a digital format. Most individuals were found to produce reliable and reproducible bands within the size range of 3,054 bp and 298 bp.
Bands larger than 3,054 bp and smaller than 298 bp were generally weak and their presence inconsistent when comparing fingerprints between replicate gels. The resulting digital profiles for a given beginner individual therefore consist of a series of ones and zeroes ranging from as many as ten digits to five, as shown in Figure 2.2. A numerical sequence of zeros and ones, called a binary profile (fingerprint), was recorded in Notepad, Windows' internal text editor.

ANALYSIS OF PRIMER PERFORMANCE
ANALYSIS OF RAPD FINGERPRINTS
- Quantification of genetic relationships
An analysis of molecular variance (AMOVA) was also performed to determine the contribution of the between and within population or group variation to the total variation. In this investigation, the proportion of polymorphism was determined for the population as a whole as well as for each of the different families. Heterozygosity can be estimated in a number of different ways, each method giving a value that indicates the genetic variation in the population.
From the results of genetic identity and genetic distance measurements, a dendogram was created showing the relationship between the four families and the population. In the case of a cross-family input file, the data were grouped into four sections representing different families and labeled accordingly. Once the data were loaded into the program, analysis of genetic variation and genetic relationship between and within families was determined by dominant marker analysis on the diploid data.

ARLEQUIN
An AMOVA analysis was performed on the data from the specified project file, constructing a Euclidean distance matrix, using 1,023 permutations. The 'Run' function was then selected, after which the specified calculations were undertaken by the program. The results were viewed in the AMOVA file as a separate file, with the extension '.res', and interpreted.
RESULTS
INTRODUCTION
ASSESSMENT OF DNA ISOLATION METHODOLOGY
The quality of the isolated DNA was also assessed by gel electrophoresis to verify whether the DNA was high molecular weight or degraded. Although three different ages of leaf samples were used in the study for an appropriate DNA isolation protocol, it was found that the concentration of DNA was more or less the same in all age groups studied. After the preliminary evaluation of the three DNA isolation methods, the CTAB method proposed by Doyle and Doyle (1990) was chosen as the most effective method for the isolation of DNA from black leaves.
However, slight optimizations of the original method were undertaken by varying the concentrations of polyvinylpolypyrrolidone (PVP) and 2-mercapto-ethanol. This purity indicated that the isolated DNA was very pure as purity values ranged within the standard range of pure DNA; 1.8 and 2.0. In this investigation, the concentrations of DNA, primer, and magnesium chloride (MgCb) of the PCR reaction mixture were optimized by varying the concentrations of a particular reagent while keeping the other reagents in the reaction mixture constant.

41 CYCLES
ANALYSIS OF RAPD FINGERPRINTS
The presence of a band indicated the dominant phenotype of genotypes AA or Aa, and was scored as a. The performance of the 14 primers used in this study was assessed for their ability to amplify polymorphic loci by evaluating all fingerprints. Primers were called polymorphic if some loci produced bands that were present in some individuals and absent in others, while primers were called monomorphic if all loci produced bands in all individuals.
In this investigation, five of the 14 primers were found to be monomorphic and therefore excluded from subsequent analyses, leaving 68 analyzable loci. To determine whether the GC content of the primers affected the number of loci amplified in a fingerprint or not, the number of amplified loci from the primers with different GC contents was compared. From Table 3.6 it became clear that no specific relationship could be established between the percentage of GC content of the primers and the number of bands they produced.

ASSESSMENT OF GENETIC VARIATION
- RAPD markers associated with gummosis
Three measures of genetic variation, polymorphic percentage, Nei's diversity and Shannon's information index, clearly showed a rather limited amount of genetic variation in this population, which was also reflected in each of the families (Table 3.7). Population heterozygosity (2pq) was assessed by first determining the frequency of the null phenotype at each locus in all comparisons. Next, the null allele frequency (q) was calculated by calculating the square root of the null phenotype frequency.
Then, the frequency of heterozygotes (2pq) was estimated for each site, after which the mean for the population was calculated (Table 3.8). It was expected that within each of the families genetic variation would be low, however, it was an unexpected result that the families were so closely related. The trees provided a representation of the overall genetic relationships between individuals within a family and between families, with branch lengths indicating an estimate of frequency differences (Figure 3.7).

I ~lndividual4 1
A comparison of the genetic relationship between the different families revealed that the three gummose-susceptible families clustered together in the tree with short arm lengths, while family 18, gummose-resistant, separated on a separate branch from these families is. A tree was constructed using TREEVI EW (Page, 1996) to relate the relationships between all the individuals of the population (Figure 3.8). A hierarchical analysis of molecular variance (AMOVA) was performed to quantify and partition the levels of variation into within- and within-form components.
The results of the AMOVA partitioning of RAPD variance between and within families are shown in Table 3.9. Of the total genetic variation, 87.86% was attributable to within-family variation and only 12.14% to inter-family variation in the black band. However, the proportion of variation attributed between families and variation within families was found to be highly significant (P < 0.001), indicating significant genetic differentiation between families and within families.

DISCUSSION AND CONCLUDING REMARKS
Isolation of DNA from black river has proven to be problematic due to the presence of high levels of polysaccharides, polyphenols and tannins present in the cells. They may also interfere with the quantitative determination of nucleic acid purity by spectrophotometric methods (Wilkie et al., 1993). In a tree improvement program, knowledge of the distribution of genetic variation within and among natural tree populations is important for initial selection.
Many studies have been conducted on the determination of genetic variation within and between populations of different species. All these measures of variation clearly indicated a rather limited amount of genetic variation in this population, which was also true for each of the families. Therefore, it is important that a large-scale investigation of genetic variabilities be carried out across the entire distribution range of black mulberry plantations to obtain a more accurate estimate of the extent and complexity of black mulberry genetic variability.
LABORATORY SOLUTION RECIPES
POPGENE ANALYSIS
ARLEQUIN ANALYSIS
APPENDIX A
DNA isolation protocol 2
DNA isolation protocol 3
All Families
Above diagonal: Average number of pairwise differences between populations (PiXY) Diagonal elements: Average number of pairwise differences within population (PiX) Below diagonal: Corrected average pairwise difference (PiXY-(PiX+PiY)j2). Allowed level of missing data Number of indels observed Number of polymorphic sites Number of usable nucleotide sites. Allowed level of missing data Number of indels observed Number of polymorphic sites Number of usable nucleotide sites.




