Assessing Biological Impacts of Land Reclamation in a Mining
Region in Canada: Effects of Dolomitic Lime Applications
on Forest Ecosystems and Microbial Phospholipid Fatty Acid
Signatures
K. K. Nkongolo&P. Michael&G. Theriault&R. Narendrula& P. Castilloux&K. N. Kalubi&P. Beckett&G. Spiers
Received: 13 July 2015 /Accepted: 1 March 2016 / Published online: 10 March 2016 #Springer International Publishing Switzerland 2016
Abstract The ability of an ecosystem to withstand se-rious disturbances may depend in part on the diversity of the system, with soil microbiological communities be-ing coupled to their associated vegetation. The main objectives of the present study were (1) to determine the association between soil microbial diversity, abun-dance, and activities with diversity and sustainability of remediated mining-damaged ecosystems in Northern Ontario, and (2) to assess the effects of dolomitic lime applications on aboveground and belowground biodi-versity and community structures. Results revealed that liming increases soil pH, cation exchange capacity (CEC), forest tree species diversity and abundance, and the overall ecosystem health even 25 to 35 years after dolomite applications. The mean Shannon index
value was significantly higher in limed compared to unlimed sites. Tree species richness was 4.0, 6.0, and 7.7 for unlimed, limed, and reference sites, respectively. Overall, the mean health index revealed a significant improvement in population health in limed sites (index score = 7) compared to unlimed areas (index score = 4). Soil microbial biomass and respiration were also in-creased by liming. Surprisingly, the main component of soil microbial biomass in limed, unlimed, and refer-ence sites within the vicinity of the restored lands was bacteria (mostly Gram (−). Significant difference was
also observed between limed, unlimed, and reference sites for arbuscular mycorrhizal fungi, Gram (−)
bacte-ria, Gram (+) bactebacte-ria, anaerobe, and actinomycetes abundance. The ratios between fungi and bacteria and among other phospholipid fatty acid (PLFA) measures were extremely low suggesting that the targeted region is still under environmental stress. No apparent associ-ations among soil microbial biomass, soil respiration, and forest plant diversity or abundance were observed. Soil pH levels and organic matter amounts and quality appear to be the main factors affecting these parameters. Molecular analysis of main tree species within the im-pacted region revealed no change in genetic variation among plant populations on either limed or unlimed lands contaminated with metals over two generations.
Keywords Soil liming . Plant diversity and abundance . ISSR analysis . Soil respiration . PLFA . Microbial biomass
DOI 10.1007/s11270-016-2803-5
K. K. Nkongolo (*)
:
P. BeckettDepartment of Biology, Laurentian University, Sudbury, ON P3E 2C6, Canada
e-mail: knkongolo@laurentian.ca
K. K. Nkongolo
:
P. Michael:
G. Theriault:
R. Narendrula:
K. N. KalubiDepartment of Biomolecular Sciences program, Laurentian University, Sudbury, ON P3E 2C6, Canada
P. Castilloux
Collège Boréal, 21 Lasalle Blvd, Sudbury, ON P3A 6B1, Canada
G. Spiers
1 Introduction
Soil biota is implicated in most of the key functions soil provides in terms of ecosystem services, driving many fundamental nutrient cycling processes, soil structural dynamics, degradation of pollutants, and regulation of plant communities. The protection of soil organism biodiversity is a major way to maintain the proper functioning of the soil. The living population inhabiting soil includes macrofauna, mesofauna, microfauna, and microflora (Cannon1999; Nannipieri et al.2003)
There is evidence that soil biotic communities are coupled to their associated vegetation, with a mutual dependence between aboveground and belowground communities such that compromised soil communities may curtail formation of particular plant assemblages (Luo and Zhou2006). Recent studies suggest that, by characterizing diversity, one will be able to understand and manipulate the working of ecosystems. The ability of an ecosystem to withstand serious disturbances may depend in part on the diversity of the system (Nannipieri et al.2003).
Fungi and bacteria play crucial roles in ecosystem function. They constitute the dominant organism groups involved in C/N cycling, primarily in the degradation of organic matter. Critical stages in lignin decomposition are almost exclusively carried out by fungi, with the breakdown of complex biomolecules such as cellulose and tannins in soils being due mostly to fungal enzy-matic activity. Fungi account for up to 90 % of total living biomass in forest soils, and soil fungal respiration has been found to be two to four times greater than that of bacteria (Anderson and Domsch 1978). The long-term stability of ecosystems is therefore dependent on the continued contribution of fungal activities. Little is known about the structure and composition of the mi-crobial communities inhabiting Northern Ontario (Canada) soils, and thus of the biological processes shaping the assembly of those communities.
The Greater Sudbury region of Northern Ontario is known globally for extensive nickel, copper, and other metal deposits, with the aerosolic emissions from the roasting and smelting of the ores rich in these elements causing disastrous effects on the vegetation and overall environment (Amiro and Courtin1981; Gratton et al.
2000; Narendrula et al. 2012; Nkongolo et al. 2008; Vandeligt et al. 2011). More than 80,000 ha became semi-barren to completely barren, and several studies implicated sulfur dioxide emissions and metal
particulates in soils as being the main cause of these effects (Wren et al. 2012). Dolomitic lime which con-tains calcium and magnesium carbonate was applied to soils from 1978 to today as part of a Land Reclamation (Regreening) Programme in the Greater Sudbury region It is believed that at least some minimum number of species are essential for ecosystem functioning under steady conditions, with a large number of species prob-ably being important for maintaining stable processes in changing environments (Nannipieri et al.2003). Threats to soil systems such as erosion, decline of organic matter content, and contamination will likely compromise or destroy the soil biota and thus affect soil biodiversity. However, even polluted or severely disturbed soils still support relatively high levels of microbial diversity with bacterial populations being more susceptible to certain soluble contaminant metals than fungi (Khan2000).
The main objectives of the present study were (1) to determine the association between soil microbial diver-sity, abundance, and activities with diversity and sus-tainability of mining-damaged ecosystems in Northern Ontario, and (2) to assess the effects of applications of dolomitic liming materials on aboveground and below-ground biodiversity and community structures.
2 Materials and Methods
2.1 Sampling
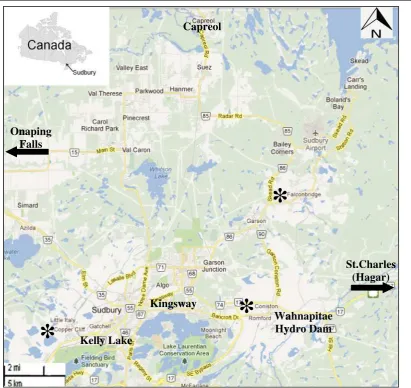
Nine forest populations in six areas were selected for this study. Three areas were contaminated with metals and each had limed and unlimed sites; three other areas were metal-uncontaminated and used as comparison sites. Contaminated sites were located within the 81,000 ha affected by smelter emissions and include Wahnapitae Hydro Dam, Kingsway Road, and Kelly Lake sites (Fig. 1). They were highly or moderately disturbed based on their proximity to smelters as de-scribed in Nkongolo et al. (2013). Comparison sites were at St. Charles (Hagar), Onaping Falls, and Capreol at distances >40 km from Sudbury.
2.2 pH and Cation Exchange Capacity
acetate (pH 7) extracts of soil samples as developed by Lavkulich1981, with the total exchange capacity being estimated as the sum of the exchangeable cations as described in Hendershot et al. (2000).
2.3 Plant Species Diversity, Abundance, and Population Health
Plant community structures were analyzed using established ecological assessment procedures (Theriault et al.2013). In each population, three quad-rats of 10-m diameter each were used to assess plant species richness and abundance. Specifically, Shannon–
Wiener index, Simpson’s index of diversity, species
richness, and species evenness were estimated. A health index was also assigned to each population using a 1 to 10 scale with the grade 1 representing a population without trees or herbaceous layer (all vegetation below 50 cm in height) and a grade of 10 representing a population with maximum complexity with no apparent disturbance.
2.4 Molecular Analysis
Leaf samples from Betula papyrifera(the most domi-nant species) and Quercus rubra were collected from both limed and unlimed sites in Northern Ontario. For each species, 70 trees representing over 50 % of the
On
F
*
naping
Falls
Kelly Lake
Kingsw
e
way
Capreol
*
*
Wa
Hy
ahnapitae
ydro Dam
St.Charles
(Hagar)
s
Fig. 1 Location of red maple (Acer rubrum) sampling sites in Northern Ontario within the Greater Sudbury region. Site 1: Kelly Lake; Site 2: Kingsway; Site 3: Wahnapitae Hydro Dam; Site 4: St.
targeted population were selected. Leaf samples were wrapped in aluminum foil, immersed in liquid nitrogen, and stored at−20 °C until DNA extraction.
2.4.1 DNA Extraction
Genomic DNA was extracted from fresh frozen leaf material using the CTAB extraction protocol (Nkongolo et al.2005; Mehes et al.2007). This protocol is a modification of Doyle and Doyle (1987) procedure. The modifications included the addition of 1 % polyvi-nyl pyrrolidone (PVP) and 0.2 % beta mercaptanol to the cetyl trimethylammonium bromide (CTAB) buffer solution, two additional chloroform spins prior to the isopropanol spin, and no addition of RNAse. After extraction, DNA was stored in a freezer at−20 °C.
2.4.2 ISSR Analysis
A total of 15 inter simple sequence repeat (ISSR) primers were pre-screened for polymorphism and repro-ducibility. Of these, eight primers were identified. These are 178 99A, 178 99B, UBC 841, UBC 825, ISSR 5, 8, 9, and 10. Five of these eight primers (178 98B, UBC 825, ISSR 5, 9, and 10) that produced strong bands were selected for ISSR analysis.
PCR amplification was carried out as described by Mehes et al. (2007) in a 25-μL total volume containing a
master mix of 11.4μL distilled water, 2.5μL MgSO4, 2.1μL 10× buffer, 0.5μL of dNTPs (equal parts dTTP,
dATP, dCTP, dGTP), 0.5μL of ISSR primer, aTaqmix
of 3.475 μL distilled water, 0.4 μL 10× buffer and
0.125 μL Taqpolymerase (Applied Biosystems), and
4μL standardized DNA. Each primer contained a
neg-ative control of master mix and Taq mix without any DNA. All samples were covered with one drop of mineral oil to prevent evaporation and amplified with the Eppendorf Mastercycler gradient thermocycler. The program was set to a hot start of 5 min at 95 °C followed by 2 min at 85 °C to which theTaqmix was added. In total, 42 cycles of 1.5 min at 95 °C, 2 min at 55 °C, and 1 min of 72 °C were performed. This was followed by a final extension of 7 min at 72 °C after which samples were removed from the thermocycler and placed in a freezer set at−20 °C until further analysis.
Amplified DNA products were separated for analysis on a 2 % agarose gel in 0.5× TBE with ethidium bro-mide. Then, 5μL of 2× loading buffer was added to the
PCR products and 10μL of this solution was loaded
into the wells of the gel. The gel was run at 3.37 V/cm, documented with the Bio-Rad ChemiDoc XRS system, and analyzed with Image Lab Software. The ISSR bands on each gel were scored as either present (1) or absent (0). Popgene software version 1.32 (Yeh and Boyle
1997) was used to determine percentage of polymorphic loci, observed and effective number of alleles, Nei’s gene diversity, and Shannon’s information index. The
genetic distances were calculated using Jaccard’s
simi-larity coefficients with Free Tree Program version 1.50. A neighbor-joining dendogram was produced from the similarity coefficients. The method starts with a starlike tree with no hierarchical structure and in a stepwise fashion finds the two operational taxonomic units that minimize the total branch length at each cycle of clus-tering. The unrooted tree generated by the neighbor-joining method is constructed under the principle of minimum evolution (Saitou and Nei1987).
2.5 Microbial Analysis
2.5.1 PLFA Analysis and Soil Respiration
Phospholipid fatty acid (PLFA) analysis was performed at FAME Lab, Microbial ID. Inc, Newark, DE (USA), as described in Buyer and Sasser (2012). The procedure was originally developed by MIDI Inc. (Newark, DE) for analysis of fatty acids from laboratory- grown pure cultures. Extraction of fatty acids from the soil involves saponification of the soil at 100 °C followed by acid methylation at 80 °C for fatty acid methyl ester (FAME) formation. This was followed by an alkaline wash and extraction of FAME into hexane for analysis by gas chromatography (GC) (Kaur et al.2005).
Respiration for dried soil samples incubated for 24 h at 50 % H2O (v/v) was assessed using the Solvita Soil Test™as described in Goupil and Nkongolo (2014). A
colorimetric CO2probe inserted into the incubation jar was read using a Digital Color Reader (DCR), with interpretations of the DCR providing soil respiration data estimates based on Solvita’s guidelines.
2.6 Statistical Analysis
3 Results
3.1 Soil Acidity and CEC
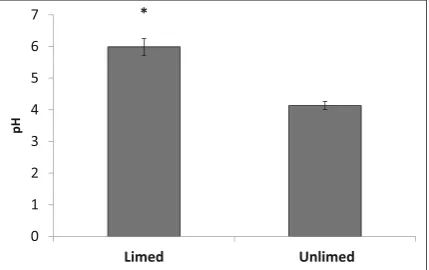
Soil pH and cation exchange capacity (CEC) estimates were significantly higher in limed areas compared to those in unlimed sites (Figs. 2 and 3). The mean pH values were 6 and 4.1 for limed and unlimed sites, respectively. CECs were 21 cmol/kg for limed sites and 2.3 cmol/kg for unlimed.
3.2 Species Richness, Abundance, and Forest Population Health
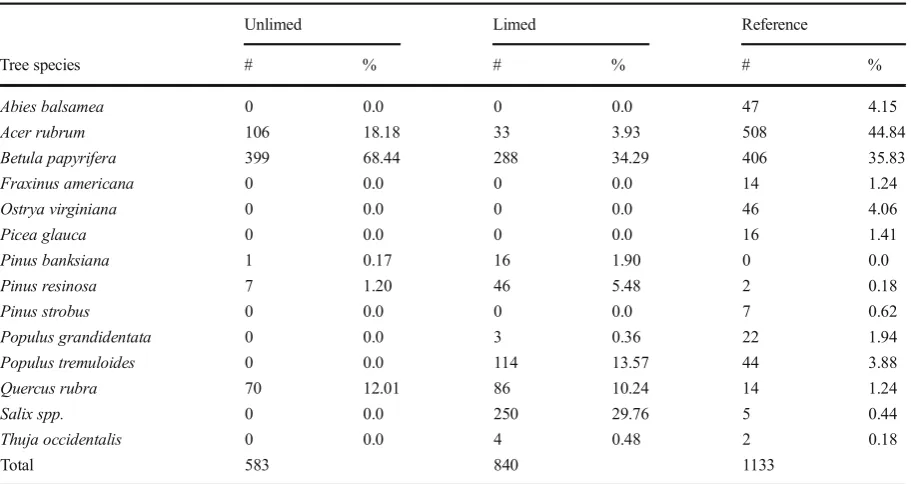
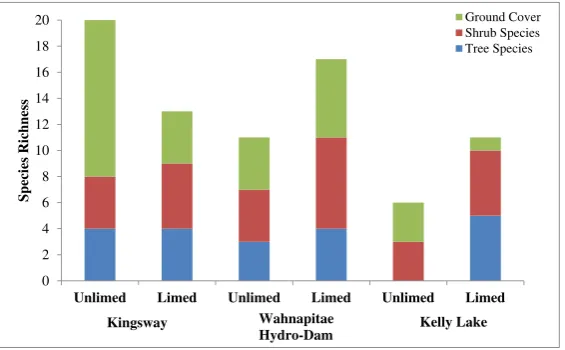
Tables1and2describe the proportions of different tree species and cover found in limed, unlimed, and refer-ence areas, with key differrefer-ences in species distribution being observed between limed and unlimed areas. In assessing species composition,B. papyriferawas more abundant in unlimed sites (68 %) than in limed (34 %). Likewise, there was a decrease ofAcer rubrumin limed sites compared to unlimed areas.Salixspp. were more prevalent in limed areas (30 %) compared to those in unlimed sections (0 %). In reference sites, A. rubrum
was the most dominant (44 %) followed by
B. papyrifera(35 %; Table1).
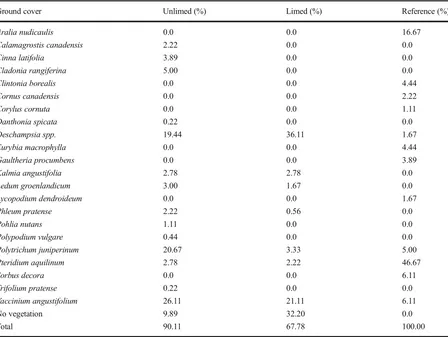
For the ground cover vegetation,Deschampsiaspp. cover was 36 and 19 % in limed and unlimed areas, respectively, followed in relative abundance by
Vaccinium angustifoliumwith 26 % in the unlimed area and 21 % in the limed areas. Polytrichum juniperum
mostly found in unlimed sites (21 %) was rare in limed areas (3 %). The ground cover in reference sites, on the
other hand, was dominated by Pteridium aquilinum
(44 %; Table2).
The mean Shannon index value was significantly higher (1.06) in limed compared to that in unlimed sites (0.76). Tree species richness was, on average, 4.0, 6.0, and 7.7 for unlimed, limed, and reference sites, respec-tively (Table 3). Figures4 and 5depict differences in tree species diversity and abundance between limed and unlimed areas. The proportion of cover was higher in unlimed sites compared to that in limed. Overall, the mean health index revealed a significant improvement in population health in limed sites compared to that in unlimed areas. The average health index values were 4, 7, and 10 for unlimed, limed, and comparison sites, respectively.
3.3 Molecular Analysis
Ten ISSR oligonucleotides primers (Table4) were used for the amplification ofQ. rubraandB. papyriferaDNA
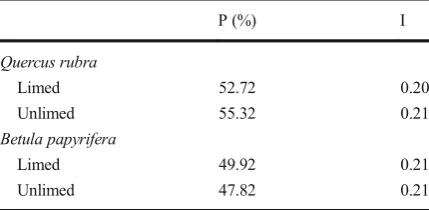
to determine the level of molecular variation since these species represent over 50 % of all tree species in the region. For each population in each species, the levels of polymorphism for the two generations analyzed were similar. Thus, the data for two generations for each species at each site were compiled and used to compare limed and unlimed populations, with no significant dif-ferences in levels of polymorphism loci being observed between limed and unlimed sites. The genetic variation was moderate in the two targeted species. Specifically, the levels of genetic variation in all populations were 54 % inQ. rubraand 49 % inB. papyriferapopulations. The mean Shannon’s information index was 0.21 in *
0 1 2 3 4 5 6 7
Limed Unlimed
pH
Fig. 2 Soil pH for limed and unlimed sites in the Greater Sudbury Region. Means were significantly different based on the Student’s
ttest (P≤0.05).Barsrepresent standard error
*
0 5 10 15 20 25
Limed Unlimed
Mean CaƟon Exchange (cmol/kg)
Fig. 3 Mean estimated cation exchange capacity (CEC) for limed
and unlimed sites in the Greater Sudbury Region. Means were significantly different based on the Student’sttest (P≤0.05).Bars
populations from both species. No significant difference for the level of polymorphism was observed between the two groups (Table5).
3.4 PLFA Analysis and Soil Respiration
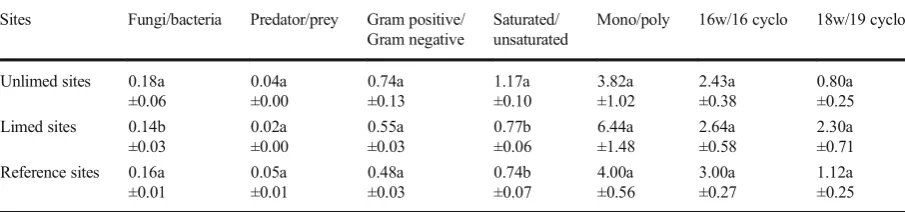
Results of phospholipid fatty acid analysis are described in Tables6and7. This analysis revealed higher micro-bial biomass in samples from limed areas compared to that in unlimed samples. Specifically, significant differ-ence was also observed among limed, unlimed, and reference sites for arbuscular mycorrhizal fungi, Gram (−) bacteria, Gram (+) bacteria, anaerobe, and
actino-mycetes abundance. Surprisingly, there were more bac-teria than fungi in all the sites analyzed (Table6). In fact, total bacteria represent 68.4, 74.3, and 71.4 % of the total microbial biomass for unlimed, limed, and refer-ence sites, respectively. These values were only 9.8 % (for unlimed), 6.7 % (for limed), and 9.4 % (for refer-ence sites) for total fungi biomass. There were twice as many Gram (−) bacteria than Gram (+) in all the sites.
The ratio between fungi and bacteria was very low for all the groups, with a significant difference being ob-served for the fungi/bacteria ratio for limed and unlimed sites (Table7).
In addition, sensible differences between limed and unlimed sites were observed for the total fungi and eukaryotes (Table 6). Surprisingly, there were more unsaturated PLFAs than saturated in limed and reference sites compared to those in unlimed samples. Overall, the ratios between unsaturated and saturated PLFA were low.
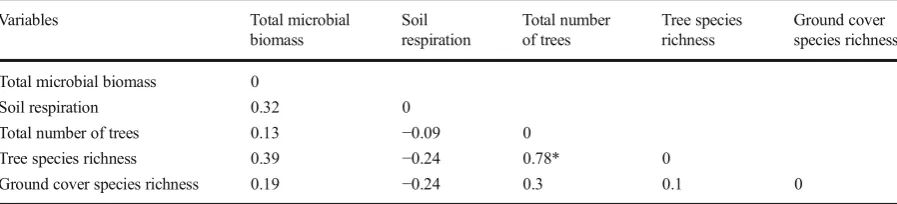
Soil respiration rates were higher in limed sites com-pared to those in unlimed areas, with the samples having been collected in the fall (Fig.6). In general, respiration rates for the reference sites were similar to those docu-mented for the limed sites, with soil bacteria, representing 70 % of the microbial communities, being the main components involved in soil respiration in these sites. No associations among soil respiration data, total soil microbial biomass, and aboveground diversity indices were observed (Table8).
4 Discussion
4.1 Soil Acidity and Metal Content
The pH values in unlimed and comparison sites were <4, consistent with acidity levels documented for soils
Table 1 Total number (#) and percentage (%) of tree species in limed, unlimed, and reference sites in Northern Ontario
Unlimed Limed Reference
Tree species # % # % # %
Abies balsamea 0 0.0 0 0.0 47 4.15
Acer rubrum 106 18.18 33 3.93 508 44.84
Betula papyrifera 399 68.44 288 34.29 406 35.83
Fraxinus americana 0 0.0 0 0.0 14 1.24
Ostrya virginiana 0 0.0 0 0.0 46 4.06
Picea glauca 0 0.0 0 0.0 16 1.41
Pinus banksiana 1 0.17 16 1.90 0 0.0
Pinus resinosa 7 1.20 46 5.48 2 0.18
Pinus strobus 0 0.0 0 0.0 7 0.62
Populus grandidentata 0 0.0 3 0.36 22 1.94
Populus tremuloides 0 0.0 114 13.57 44 3.88
Quercus rubra 70 12.01 86 10.24 14 1.24
Salix spp. 0 0.0 250 29.76 5 0.44
Thuja occidentalis 0 0.0 4 0.48 2 0.18
Total 583 840 1133
formed on coarser-textured soils with coniferous vege-tation on the Canadian Shield classified (McKeague et al. 1979; Nkongolo et al. 2013; Theriault et al.
2013; Tran et al. 2014). The pH in limed sites was significantly higher (up to 6.75) in the top organic layer (Nkongolo et al.2013). It was generally above the target
Table 2 Percentage of ground cover for different plant species in limed, unlimed, and reference sites in Northern Ontario
Ground cover Unlimed (%) Limed (%) Reference (%)
Aralia nudicaulis 0.0 0.0 16.67
Calamagrostis canadensis 2.22 0.0 0.0
Cinna latifolia 3.89 0.0 0.0
Cladonia rangiferina 5.00 0.0 0.0
Clintonia borealis 0.0 0.0 4.44
Cornus canadensis 0.0 0.0 2.22
Corylus cornuta 0.0 0.0 1.11
Danthonia spicata 0.22 0.0 0.0
Deschampsia spp. 19.44 36.11 1.67
Eurybia macrophylla 0.0 0.0 4.44
Gaultheria procumbens 0.0 0.0 3.89
Kalmia angustifolia 2.78 2.78 0.0
Ledum groenlandicum 3.00 1.67 0.0
Lycopodium dendroideum 0.0 0.0 1.67
Phleum pratense 2.22 0.56 0.0
Pohlia nutans 1.11 0.0 0.0
Polypodium vulgare 0.44 0.0 0.0
Polytrichum juniperinum 20.67 3.33 5.00
Pteridium aquilinum 2.78 2.22 46.67
Sorbus decora 0.0 0.0 6.11
Trifolium pratense 0.22 0.0 0.0
Vaccinium angustifolium 26.11 21.11 6.11
No vegetation 9.89 32.20 0.0
Total 90.11 67.78 100.00
Unlimed and limed sites: Wahnapitae Hydro Dam, Kingsway, and Kelly Lake; reference sites: St. Charles, Onaping Falls, and Capreol
Table 3 Shannon index, Simpsom’s index of diversity, species richness and evenness, and population health in limed, unlimed, and reference populations from the Greater Sudbury region, Canada
Shannon–Wiener index Simpson’s index of diversity Species richness Species evenness Population health
Tree species
Unlimed 0.69a 0.63b 3.67a 0.53a 4.00
Limed 1.25b 0.35a 6.67b 0.55a 7.67
Reference 0.97ab 0.54b 7.67b 0.50a 10.00
Ground cover
Unlimed 1.30b 0.32a 6.33b 0.75b 4.00
Limed 0.79a 0.38a 3.67a 0.45a 7.67
Reference 1.39b 0.32a 6.00b 0.78b 10.00
pH of 5 of the Sudbury Land Reclamation Program. The CEC followed the same trend with values higher in limed compared to those in unlimed sites.
Previous reports describing metal content of the same sites revealed that the limed samples contained higher levels of total and bioavailable Ca and Mg compared to those in unlimed sites (Nkongolo et al.2013; Theriault et al.2013). There were also higher levels of total and bioavailable P in limed samples compared to those in unlimed. On the other hand, higher contents of bioavail-able Al, Co, Fe, Ni, Sr, and Zn were observed in unlimed sites compared to those in limed areas (Nkongolo et al.
2013; Theriault et al. 2013), reflective perhaps of the slightly increased solubility of metal-hosting minerals at lower pH.
4.2 Ecological Analysis
Among the various indices used in ecological diversity assessment, the Shannon–Wiener index is the most
widely used for comparing diversity among various habitats (Clarke and Warwick2001; Ma2005; Stirling and Wilsey2001; Wilsey and Stirling2007). Shannon’s
index is an entropy measure and has mathematical prop-erties useful for combining diversity measures, e.g., when calculating regional diversity. In the present study, Shannon–Wiener and Simpson indices were used to compare limed and unlimed species. The Shannon–
Wiener index is more sensitive to richness and less sensitive to evenness compared to the Simpson diversity index (Clarke and Warwick2001; Ma2005; Stirling and Wilsey2001; Wilsey and Stirling2007). The two indi-ces are more robust than species richness—the number of species in the populations. Diversity and evenness can be positively or negatively related, and evenness and diversity indices are not consistently regulated by richness (Ma 2005). In general, in both limed and unlimed sites, the diversity indices were low but the diversity and evenness in limed sites were higher than in unlimed areas. As in previous analysis (Theriault
0 200 400 600 800 1000 1200
Unlimed sites Limed sites Reference sites
Total number of trees
Fig. 4 Total number of trees for unlimed, limed, and control sites from the Greater Sudbury Region. Unlimed and limed sites: Wahnapitae Hydro Dam, Kingsway, and Kelly Lake; reference sites: St. Charles, Onaping Falls, and Capreol
0 2 4 6 8 10 12 14 16 18 20
Unlimed Limed Unlimed Limed Unlimed Limed
Species Richness
Ground Cover Shrub Species Tree Species
Wahnapitae
Hydro-Dam Kelly Lake Kingsway
et al.2013), the distributions of individual trees in the populations were uneven on all the sites. The reference sites also showed a moderate level of diversity that was nevertheless significantly higher than in limed and unlimed sites. The most significant differences between limed, unlimed, and reference sites were observed in forest population health. There was a significant improvement of population growth quality in limed compared to that in unlimed sites. But, there is still a gap between limed areas and comparison sites based on forest population health.
It should be pointed out that the population assess-ment used in the present study is based on the Aubin (2012) and Drenovsky et al. (2012) approach with mod-ifications. The current rating is based on tree canopy and vegetation damages, together with the overall forest
density. This adjustment is more appropriate for assessing site recovery following damages caused by air pollutants. The scale of 1 to 10 was easy to apply since both extremes that include severely damaged sites (with no trees and limited or no vegetation) and highly dense forests (used as reference sites) were present in the targeted region. When changes of forest ecosystem are induced by human, they may lead to a global loss of resilience and the increase of vulnerability of ecosys-tems to future disturbance (Aubin2012; Laliberte et al.
2010).
4.3 Molecular Analysis
Most of the forest ecosystems within the Greater Sud-bury area of Northern Ontario have been improved considerably during the last 30 years (Dudka et al.
1996; Gratton et al.2000; Nkongolo et al.2013) through tree planting which followed significant reductions in smelter emissions. To assess forest population sus-tainability, biodiversity and genetic diversity are the key parameters considered for ecosystem stability (Mehes-Smith et al. 2009; Rajora and Mosseler
2001a, b). Losses of rare alleles, lower heterozygoty, and directional selection have been concerns noted for plant populations growing in stressed environ-ment (Slatkin 1985).
Several molecular markers have been used to deter-mine genetic variability in different taxa. The inter-simple sequence repeat (ISSR) system is a very useful and efficient tool used in assessing genetic variation of plant populations, combining features of SSRs and am-plified fragment length polymorphism (AFLP) to the universality of random polymorphic DNA (RAPD) (Pradeep Reddy et al. 2002). There are a number of advantages associated with the ISSR multi-locus tech-nique. First, ISSRs are universal in the sense that micro-satellite repeats are found in every eukaryotic genome studied to date. Second, unlike their RAPD counter-parts, ISSRs have high reproducibility. This reproduc-ibility is most likely due to the longer lengths of the primers which permit the use of higher annealing tem-peratures which, in turn, reduce non-specific binding and results in higher stringency (Bornet and Branchard
2004; Qian et al. 2001). Furthermore, contrary to microsatellites, amplification does not require prior knowledge of the DNA sequence. ISSRs are quick and easy to handle and reveal multi-locus, highly polymor-phic patterns. Finally, because each band corresponds to
Table 4 ISSR primers used for amplification of DNA from
Quercus rubra(red oak) andBetula papyrifera(white birch) tree
samples from limed, unlimed, and reference sites in Northern Ontario
Primer identification
Nucleotide sequence (5′→3′) G + C content (%)
ISSR 5 ACG ACG ACG ACG AC 64.29 ISSR 9 GAT CGA TCG ATC GC 57.14 ISSR 10 CTT CTT CTT CTT CTT CCT CCT
CCT CCT CCT CCT CT
51.43
17899A CAC ACA CAC ACA AG 50.00 17898B CAC ACA CAC ACA GT 50.00 UBC 825 ACA CAC ACA CAC ACA CT 47.06 UBC 827 ACA CAC ACA CAC ACA CG 52.94 UBC 841 GAA GGA GAG AGA GAG AYC 50.00
Table 5 Levels of polymorphism and Shannon’s information
index inQuercus rubra(red oak) andBetula papyrifera(white birch) populations from the Greater Sudbury region based on ISSR data
P (%) I
Quercus rubra
Limed 52.72 0.20
Unlimed 55.32 0.21
Betula papyrifera
Limed 49.92 0.21
Unlimed 47.82 0.21
a DNA sequence delimited by two inverted microsatellites, the amplified products, usually 200–
2000 bp long, are detectable by both agarose and poly-acrylamide gel electrophoresis (Pradeep Reddy et al.
2002).
In the present study, molecular analysis of white birch (B. papyrifera) and red oak (Q. rubra) that repre-sent over 50 % of all the tree populations in the targeted areas was completed. Molecular analysis showed that the average level of polymorphic loci was 54 and 49 % for Q. rubra and B. papyrifera, respectively. These values indicate a moderate level of genetic variability, with no significant differences in polymorphism being observed among populations from limed and unlimed areas. This observation suggests that, although the level of tree population diversity and abundance has increased in limed areas, the genetic variability within each spe-cies remains unchanged over two generations. There were also no differences between metal-contaminated and metal-uncontaminated populations at the reference sites for genetic variability, a result consistent with pre-vious data on conifer species (Dobrzeniecka et al.2011;
Narendrula et al.2013; Vandeligt et al.2011). This result suggests that the level of metals in plant tissue is too low to play an important role in genotypic selection and changes of the allelic frequencies.
4.4 Microbial Ecology
Microorganisms respond differently to prevailing envi-ronmental conditions, with different forest soil and veg-etation properties influencing the composition of the soil microbial community in a specific way (Hackl et al.
2005). In the present study, PLFA was used to analyze the composition of the microbial communities of limed, unlimed, and reference lands. The resulting data re-vealed that liming conditions increase forest diversity, complexity, and health, with the PFLA data also highlighting increases of total microbial biomass, arbuscular mycorrhizal (AM) fungi, total fungi, Gram (−) and Gram (+) bacteria, eukaryotes, anaerobe, and
actinomycetes in limed sites compared to unlimed areas. Forest soil PLFA patterns have been found to change in response to liming, application of wood ash, and fire
Table 6 Total microbial biomass, AM fungi, fungi, bacteria, eukaryote, anaerobe, and actinomycetes in soil samples from the Greater Sudbury Region using phospholipid fatty acid (PLFA) analysis
Sites Total microbial biomass AM fungi Fungi Gram negative Gram positive Eukaryote Anaerobe Actinomycetes
Unlimed sites 145.72a
Means in columns with a common subscript are not significantly different based on Tukey multiple comparison test (P> 0.05). Unlimed and limed sites: Dam, Kingsway, and Kelly Lake; reference sites: Onaping Falls, Capreol, and Hagar. Data in ng/g
Table 7 Phospholipid fatty acid (PLFA) ratios analyzed in soil samples from limed, unlimed, and reference sites in Northern Ontario
Sites Fungi/bacteria Predator/prey Gram positive/ Gram negative
Saturated/ unsaturated
Mono/poly 16w/16 cyclo 18w/19 cyclo
Unlimed sites 0.18a
(Baath et al.1995; Frostegard et al.1993; Hackl et al.
2005). Soil pH appears to be responsible for the differ-entiation in soil microbial communities rather than total metal contamination. Frostegard et al. (1996) found an increase in fungal biomarkers with different levels of soluble metal contamination (except Cu) in laboratory experiments, an aspect not observed in the present study because the level of bioavailable metals in soil solutions was too low to generate any biomarkers. Soil chemical composition and physical structure play also a critical role in PFLA profile, with the low pH reference sites and the less acidic limed sites showing a similar PLFA profile. The high level of total and AM fungi in reference and limed sites compared to that in unlimed areas could be also the result of the higher organic matter content observed in these sites as demonstrated by Baath et al. (1995) and Bardgett et al. (2001). Sur-prisingly, the overall microbial communities in all the sites were dominated by bacteria, mostly Gram (−),
indicating that the entire targeted region is still under severe environmental stress.
4.5 Relation Between Microbial Biomass, Activities, and Aboveground Diversity
The present study revealed no correlation between total microbial biomass in the soil and forest complexity and diversity. These two components are, however, func-tionally linked since microbes facilitate mineralization of organic matter for both plant nutrient supply and the provision of an appropriate physical environment for microbial growth. The lack of association between soil microbial biomass, soil respiration, and plant population diversity could be, in part, attributed to the severe level of land damage and the soil acidity level. The conse-quence of limited soil biodiversity is not well under-stood. From a functional perspective, soil species rich-ness might be of little consequence, but the functional repertoire of the soil biota is critical. There are several microbial species involved in organic matter decompo-sition. But, processes such as nitrification (the oxidation of ammonium) are carried out by a narrower range of bacteria and there is less redundancy in this group except for highly specific symbiotic associations. Re-duced biodiversity will have differing consequences in relation to different soil activities, and some specific soil processes are impaired below a certain level of soil biodiversity. A metagenomic analysis of soil sample is in progress to establish the types of fungi and bacteria present in each site and the level of soil microbial diversity.
5 Conclusions
The ability of an ecosystem to withstand extreme dis-turbance may depend in part on the biotic and functional diversity of the system. The latter is a function of species 0
10 20 30 40 50 60 70 80
Limed Unlimed Control
Soil RespiraƟon CO2-C(mg/kg
(ppm))
Limed Unlimed Control
b
a a
Fig. 6 Fall soil respiration rates for unlimed, limed, and reference
sites in the Greater Sudbury Region. Means with a commonletter
are not significantly different based on Tukey multiple comparison test (P> 0.05)
Table 8 Correlation between microbial biomass, soil respiration, and ecological parameters for the unlimed, limed, and reference sites from Northern Ontario
Variables Total microbial
biomass
Soil respiration
Total number of trees
Tree species richness
Ground cover species richness
Total microbial biomass 0
Soil respiration 0.32 0
Total number of trees 0.13 −0.09 0
Tree species richness 0.39 −0.24 0.78* 0
Ground cover species richness 0.19 −0.24 0.3 0.1 0
richness or species abundance and species evenness or species equitability. In the present study, these parame-ters were measured to assess the sustainability of dam-aged land following restoration mainly through the ap-plication of dolomitic limestone. Results from different analyses revealed that liming increases forest trees di-versity and abundance and the overall ecosystem health even 25 to 35 years after dolomitic applications. Soil microbial biomass and respiration were also increased in soils following lime application. The main component of soil microbiome in limed, unlimed, and comparison sites within the vicinity of the reclaimed lands was Gram (−) bacteria. The ratios between fungi and bacteria and
among other PLFA measures were extremely low sug-gesting that the targeted region is still under environ-mental stress. No associations among soil microbial biomass, soil respiration, and forest plant diversity or abundance were observed. Soil pH and organic matter appear to be the main factors affecting these parameters. However, molecular analysis of main tree species within the targeted region revealed no change in genetic vari-ation among plant populvari-ations from limed and unlimed lands contaminated with metals over two generations.
Acknowledgments We would like to thank the Natural Sciences and Engineering Research Council of Canada (NSERC), Vale (Sudbury), and Sudbury Integrated Nickel Operations - Glencore Company (formerly Xstrata Limited) for their financial support.
References
Amiro, B. D., & Courtin, G. M. (1981). Patterns of vegetation in the vicinity of an industrially disturbed ecosystem Sudbury Ontario Canada.Canadian Journal of Botany, 59(9), 1623–
1639.
Anderson, J. P. E., & Domsch, K. H. (1978). A physiological method for the quantitative measurement of microbial bio-mass in soils.Soil Biology and Biochemistry, 10(3), 215–221. Aubin, I. (2012). From seed size to ecosystem health: the plant trait
approach.Frontline Express, 57(57).
Baath, E., Frostegard, A., Pennanen, T., & Fritze, H. (1995). Microbial community structure and pH response in relation to soil organic matter quality in wood-ash fertilized, clear-cut or burned coniferous forest soils. Soil Biology and
Biochemistry, 27(2), 229–240.
Bardgett, R., Jones, A., Jones, D., Kemmitt, S., Cook, R., & Hobbs, P. (2001). Soil microbial community patterns related to the history and intensity of grazing in sub-montane eco-systems.Soil Biology and Biochemistry, 33(12–13), 1653–
1664.
Bornet, B., & Branchard, M. (2004). Use of ISSR fingerprints to detect microsatellites and genetic diversity in several related
Brassica taxaandArabidopsis thaliana. Hereditas, 140(3),
245–248.
Buyer, J. S., & Sasser, M. (2012). High throughput phospholipid fatty acid analysis of soils.Applied Soil Ecology, 61, 127–
130.
Cannon, P. F. (1999). Options and constraints in rapid diversity analysis of fungi in natural ecosystems.Fungal Diversity, 2, 1–15.
Carter, M. R. (1993).Soil sampling and methods of analysis. Boca Raton: Lewis.
Clarke, R. K., & Warwick, R. (2001). A further biodiversity index applicable to species lists: variation in taxonomic distinct-ness.Marine Ecology Progress Series, 216, 265–278. Dobrzeniecka, S., Nkongolo, K. K., Michael, P., Mehes-Smith,
M., & Beckett, P. (2011). Genetic analysis of black spruce
(Picea Mariana) populations from dry and wet areas of a
metal-contaminated region in Ontario (Canada).Water, Air,
and Soil Pollution, 215(1–4), 117–125.
Doyle, J. J., & Doyle, J. L. (1987). A rapid DNA isolation procedure for small quantities of fresh leaf tissue.
Phytochemical Bulletin, 19, 11–15.
Drenovsky, R. E., Grewell, B. J., D'Antonio, C. M., Funk, J. L., James, J. J., Molinari, N., Parker, I. M., & Richards, C. L. (2012). A functional trait perspective on plant invasion.
Annals of Botany, 110(1), 141–153.
Dudka, S., Ponce-Hernandez, R., Tate, G., & Hutchinson, T. C. (1996). Forms of Cu, Ni and Zn in soil of Sudbury, Ontario and the metal concentrations in plants.Water, Air, and Soil
Pollution, 90(3–4), 531–542.
Frostegard, A., Baath, E., & Tunlid, A. (1993). Shifts in the structure of soil microbial communities in limed forests as revealed by phospholipid fatty acid analysis.Soil Biology
and Biochemistry, 28(1), 55–63.
Frostegard, A., Tunlid, A., & Baath, E. (1996). Changes in micro-bial community structure during long-term incubation in two soils experimentally contaminated with metals.Soil Biology
and Biochemistry, 25(6), 723–730.
Goupil, K., & Nkongolo, K. (2014). Assessing soil respiration as an indicator of soil microbial activity in reclaimed metal contaminated lands.American Journal of Environmental
Sciences, 10, 403–411.
Gratton, W., Nkongolo, K., & Spiers, G. (2000). Heavy metal accumulation in soil and jack pine (Pinus banksiana) needles in Sudbury, Ontario, Canada.Bulletin of Environmental
Contamination and Toxicology, 64(4), 550–557.
Hackl, E., Pfeffer, M., Donat, C., Bachmann, G., & Zechmeister-Boltenstern, S. (2005). Composition of the microbial com-munities in the mineral soil under different types of natural forest.Soil Biology and Biochemistry, 37(4), 661–671. Hendershot, W. H., Lalande, H., & Duquette, M. (2000). Ion
exchange and exchangeable cations. In M. R. Carter & E. G. Gregorich (Eds.),Soil sampling and methods of analysis
(2nd ed).
Kaur, A., Chaudhary, A., Kaur, A., Choudhary, R., & Kaushik, R. (2005). Phospholipid fatty acid—a bioindicator of environ-ment monitoring and assessenviron-ment in soil ecosystem.Current
Science (Bangalore), 89(7), 1103–1112.
Khan, M. (2000). Effect of metals contamination on soil microbial diversity, enzymatic activity, organic matter decomposition and nitrogen mineralization (a review).Pakistan Journal of
Laliberte, E., Wells, J. A., DeClerck, F., Metcalfe, D. J., Catterall, C. P., Queiroz, C., Aubin, I., Bonser, S. P., Ding, Y., Fraterrigo, J. M., McNamara, S., Morgan, J. W., Sanchez Merlos, D., Vesk, P. A., & Mayfield, M. M. (2010). Land-use intensification reduces functional redundancy and response diversity in plant communities.Ecology Letters, 13(1), 76–
86.
Lavkulich, L. J. (1981). Methods manual. Vancouver, BC: Pedology Laboratory, Department of Soil Science, University of British Columbia.
Luo, Y., & Zhou, X. (2006).Soil respiration and the environment. Burlington: Academic Press.
Ma, M. (2005). Species richness vs evenness: independent rela-tionship and different responses to edaphic factors.Oikos, 111(1), 192–198.
McKeague, J. A., Desjardins, J. G., & Wolynetz, M. S. (1979). Minor elements in Canadian soils.Agriculture Canada, Research Branch Land Resource Research Institute
Contribution No LRRI 27.
Mehes, M. S., Nkongolo, K. K., & Michael, P. (2007). Genetic analysis ofPinus strobusandPinus monticolapopulations from Canada using ISSR and RAPD markers: development of genome-specific SCAR markers.Plant Systematics and
Evolution, 267(1–4), 47–63.
Mehes-Smith, M., Nkongolo, K., & Kim, N. S. (2009). A com-parative cytogenetic analysis of five pine species from North America,Pinus banksiana, P. contorta, P. monticola,
P. resinosa, and P. strobus. Plant Systematics and
Evolution, 292, 153–164.
Nannipieri, P., Ascher, J., CECcherini, M. T., Landi, L., Pietramellara, G., & Renella, G. (2003). Microbial diversity and soil functions.European Journal of Soil Science, 54(4), 655–670.
Narendrula, R., Nkongolo, K., & Beckett, P. (2012). Comparative soil metal analyses in Sudbury (Ontario, Canada) and Lubumbashi (Katanga, DR-Congo). B u l l e t i n o f
Environmental Contamination and Toxicology, 88(2), 187–
192.
Narendrula, R., Nkongolo, K. K., Beckett, P., & Spiers, G. (2013). Total and bioavailable metals in two contrasting mining regions (Sudbury in Canada and Lubumbashi in DR-Congo): relation to genetic variation in plant populations.
Chemistry and Ecology, 29(2), 111–127.
Nkongolo, K. K., Michael, P., & Demers, T. (2005). Application of ISSR, RAPD, and cytological markers to the certification of
Picea mariana,P. glauca, andP. engelmanniitrees, and their
putative hybrids.Genome, 48(2), 302–311.
Nkongolo, K. K., Vaillancourt, A., Dobrzeniecka, S., Mehes, M., & Beckett, P. (2008). Metal content in soil and black spruce
(Picea mariana) trees in the Sudbury region (Ontario,
Canada): low concentration of arsenic, cadmium, and nickel detected near smelter sources.Bulletin of Environmental
Contamination and Toxicology, 80(2), 107–111.
Nkongolo, K. K., Spiers, G., Beckett, P., Narendrula, R., Theriault, G., Tran, A., & Kalubi, K. N. (2013). Long-term effects of liming on soil chemistry in stable and eroded upland areas in a mining region.Water, Air, and Soil Pollution, 224(7), 1618. Pradeep Reddy, M., Sarla, N., & Siddiq, E. A. (2002). Inter simple sequence repeat (ISSR) polymorphism and its application in plant breeding.Euphytica, 128(1), 9–17.
Qian, W., Ge, S., & Hong, D. Y. (2001). Genetic variation within and among populations of a wild riceOryza granulatafrom China detected by RAPD and ISSR markers.Theoretical and
Applied Genetics, 102(2–3), 440–449.
Rajora, O. P., & Mosseler, A. (2001a). In G. MÃller-Starck & R. Schubert (Eds.),Molecular markers in sustainable manage-ment, conservation, and restoration of forest genetic
re-sources(pp. 187–202). Netherlands: Springer.
Rajora, O., & Mosseler, A. (2001b). Challenges and opportunities for conservation of forest genetic resources.Euphytica, 118(2), 197–212.
Saitou, N., & Nei, M. (1987). The neighbor-joining method: a new method for reconstrucing phylogenetic trees.Molecular
Biology and Evolution, 4, 406–425.
Slatkin, M. (1985). Rare alleles as indicators of gene flow.
Evolution, 39, 53–65.
Stirling, G., & Wilsey, B. (2001). Empirical relationships between species richness, evenness, and proportional diversity.
American Naturalist, 158(3), 286–299.
Theriault, G., Nkongolo, K. K., Narendrula, R., & Beckett, P. (2013). Molecular and ecological characterisation of plant populations from limed and metal-contaminated sites in Northern Ontario (Canada): ISSR analysis of white birch
(Betula papyrifera) populations.Chemistry and Ecology,
29(7), 573–585.
Tran, A., Nkongolo, K. K., Mehes-Smith, M., Narendrula, R., Spiers, G., & Beckett, P. (2014). Heavy metal analysis in red oak (Quercus rubra) populations from a mining region in northern Ontario (Canada): effect of soil liming and analysis of genetic variation.American Journal of Environmental
Sciences, 10, 363–373.
Vandeligt, K. K., Nkongolo, K. K., Mehes, M., & Beckett, P. (2011). Genetic analysis of Pinus banksianaandPinus
resinosapopulations from stressed sites contaminated with
metals in Northern Ontario (Canada). Chemistry and
Ecology, 27(4), 369–380.
Wilsey, B., & Stirling, G. (2007). Species richness and evenness respond in a different manner to propagule density in devel-oping prairie microcosm communities. Plant Ecology, 190(2), 259–273.
Wren, C., Watson, G., & Butler, M. (2012).Risk assessment and environmental: a case study in Sudbury, Ontario, Canada
(1st ed.). The Netherlands: Maralte B. V. Leiden.
Yeh, F. C., & Boyle, T. (1997). Population genetic analysis of co-dominant and co-dominant markers and quantitative traits.