Atherosclerosis 154 (2001) 243 – 246
Low-density lipoprotein receptor gene (LDLR) world-wide website
in familial hypercholesterolaemia: update, new features and
mutation analysis
Karen E. Heath
a,b, Mike Gahan
c, Ros A. Whittall
a, Steve E. Humphries
a,*
aCentre for Cardio6ascular Genetics,The Rayne Institute,Uni6ersity College Medical School,5Uni6ersity Street,London WC1E6JJ,UK bUnit of Clinical Molecular Genetics,Camelia Botnar Laboratories,Great Ormond Street Hospital for Children NHS Trust,
Great Ormond Street,London WC1N3JH,UK
cInformation Systems,Kathleen Lonsdale Building,Uni6ersity College London,Gower Place,London WC1E6BT,UK
Abstract
Mutations in the low density lipoprotein receptor gene (LDLR) cause familial hypercholesterolaemia (FH). The FH website (http://www.ucl.ac.uk/fh) has been updated to provide various functions enabling the analysis of the large number of LDLR mutations. To date, 683 LDLR mutations have been reported; of these 58.9% are missense mutations, 21.1% minor rearrange-ments, 13.5% major rearrangements and 6.6% splice site mutations. Of the 402 missense mutations, only 11.4% occurred at CpG sites. The majority of mutations were found in two functional domains, the ligand binding domain (42%) and the epidermal growth factor (EGF) precursor-like domain (47%). This report describes new features of the FH website and assesses the spectrum of mutations reported to date. © 2001 Elsevier Science Ireland Ltd. All rights reserved.
Keywords:Low density lipoprotein receptor; Familial hypercholesterolaemia; Mutations
www.elsevier.com/locate/atherosclerosis
1. Introduction
Familial hypercholesterolaemia (FH) is an autosomal dominant disorder with estimated frequencies of 1 in 500 and 1 in a million in most populations for het-erozygotes and homozygotes respectively [1]. Mutations in the low-density lipoprotein receptor gene (LDLR) give rise to familial hypercholesterolaemia (FH). The gene is located on chromosome 19p13.1 – 13.3 [2], spans 45 kb and comprises of 18 exons and 17 introns that encode a mature protein of 839 amino acids [3]. The LDL-receptor is made up of six functional domains; the signal sequence, ligand binding domain, epidermal growth factor (EGF) precursor-like domain, O-linked sugar domain, transmembrane domain and the cyto-plasmic domain [3]. In 1997, a FH website for LDLR mutations was established (http://www.ucl.ac.uk/fh) [4]. Originally, the database consisted of two core elements; a gene map and a mutation list. This report describes
the updates, new features and evaluates the mutations found to date.
2. Familial hypercholesterolaemia website new and updated features
On accessing the database, the user is immediately taken to an introductory screen giving hypertext-linked access to other areas of the database, mutation database (table and gene map), major rearrangement database, polymorphism databases (description and incidence), haplotype databases, table of double mutants, table of founder gene effect mutations, tables of published LDLR oligonucleotides, and useful links and references. The mutation database continues to exist in two formats, the gene map, which has proved very useful to our laboratory, and the mutation list. The gene map is an Oracle database and mutations are represented by an asterisk with a numbering system above the mutation site. Clicking on the mutation number, displays the information on that mutation which is listed in the mutation database. The mutation list has been refor-* Corresponding author. Tel.: +44-20-7679-6969; fax: +
44-20-7679-6212; Email address: [email protected](S.E. Humphries).
K.E.Heath et al./Atherosclerosis154 (2001) 243 – 246 244
matted into a Microsoft Access database (v7.0), which enables the user to download the file and run various search and sort functions. Information on each muta-tion is provided in a variety of fields: website identifica-tion code number, nucleotide base change and number, amino acid change and number, exon location, func-tional region, direct PCR-restriction enzyme assay (if reported or submitted to the website) with loss (−) or gain (+) of site indicated, trivial name, LDL-binding activity, mutation details (nucleotides involved in minor deletions or insertions and any details on where the protein is terminated, a comments field (where any additional information is noted, for example any fre-quency data or cellular studies), ethnic origins in which the mutation has been observed, and finally authors and references. The fields are now in a format where each entry can be searched, for example all the muta-tions observed in a particular population can be listed by sorting on the ethnic origin field, or all exon 4 mutations can be listed by sorting on the exon field. Additional tables now exist, all in a Microsoft Access database format (v7.0), major rearrangements, poly-morphism description and incidence, haplotype data, double mutants and founder gene effect mutations.
The major rearrangement (size\25bp) table has at
present 92 mutations, ranging in size from a 37 bp deletion of exon 4 [5] to a\25 kb deletion of intron 11
to the 3% flanking region (FH Catania) [6]. The boundaries of the major rearrangements, exons deleted or duplicated, size, breakpoints, protein region deleted or duplicated, functional class if determined, mutation identification number code (which refers to main muta-tion database), ethnic origin, LDL-binding activity, au-thors and referees.
The polymorphism description database is a small table listing the polymorphism identification number, location, variant (marker name or the particular en-zyme that cuts one of the variants), sequence variants, nucleotide base number, original author and reference. The polymorphism incidence database has frequency data, heterozygosity (Hz) and polymorphism informa-tion content (PIC) values of the polymorphisms in normal and FH individuals from different ethnic groups, with the respective author and reference.
The haplotype database provides haplotype data re-ported on various mutations. This has been written in a format, which allows searching on particular polymor-phic variants, the various haplotypes associated with a mutation and the haplotypes found in various ethnic groups. All the double mutants reported are listed in a database with the gene location, ethnic origin, number of cases, authors and references. The mutations that are present at a higher frequency due to a founder gene effect are listed in a database with the ethnic origins, authors and references again listed.
Tables of published LDLR oligonucleotides now ex-ist, with the original two sets [1,7] also presented on the gene map. The other tables include a combination of oligonucleotides of these two sets, plus others that amplify at one annealing temperature and three sets of published oligonucleotides used in DGGE analysis [8 – 10]. The oligonucleotide tables include location, size of product and PCR and DGGE conditions.
3. Analysis of mutations
To date 683 unique mutations are found on the FH website, 592 (86.7%) involved 1 – 24 bp whilst 92 (13.5%) were major rearrangements \25 bp in size. A
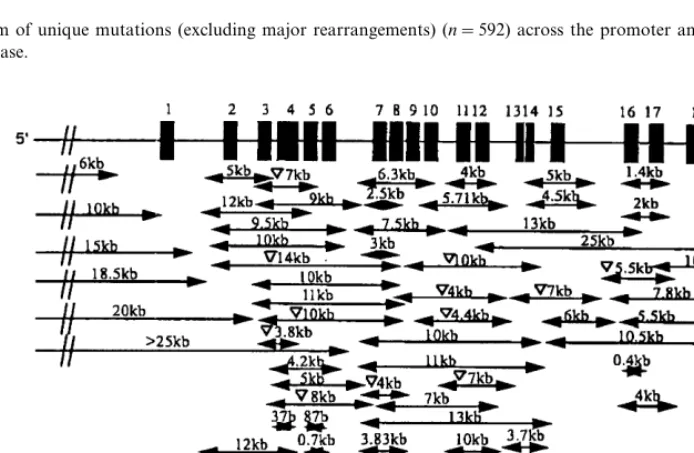
total of 45 (6.6%) splice site mutations have been reported, 144 (21.1%) minor rearrangements and 402 (58.9%) missense mutations. Only 46 of the 402 mis-sense mutations occurred at CpG sites resulting in a frequency of 11.4%, similar to that reported in the study of 157 FH homozygotes and 13 heterozygotes, 15% [1]. The spectrum of mutations (excluding major rearrangements) is shown in Fig. 1, showing the high frequency of exon 4 mutations, in particular in repeat 5, that has been associated with a more severe phenotype [11]. Mutations in the ligand binding domain and the EGF precursor-like domain accounted for 42 and 47% of mutations respectively whilst only 3.4% of mutations were found in the cytoplasmic tail (3.2% exon 17 and 0.2% exon 18), 1.7% in the membrane-spanning domain and promoter and 1.4% in the O-linked sugars domain. As shown in Fig. 2, the major rearrangements are located throughout the gene although breakpoints are more common in introns 1 – 8 and introns 12 – 3% un-translated region, where Alu repeat sequences are found at a higher frequency [12].
4. Conclusion
New mutations can still be submitted to the FH website electronically, but now mutations not seen pre-viously in a particular ethnic group and haplotype data can also be submitted. The information submitted is held until it can be reviewed appropriately and then the new mutations are added to the database. The inclusion of an e-mail address is essential for any queries and acceptance of the mutation, after review, to the database.
K.E.Heath et al./Atherosclerosis154 (2001) 243 – 246 245
systems. Finally, notification of omissions and errors in the current version and any ideas for improvement of this website would be gratefully received by the data-base curators, (e-mails found on the website acknowl-edgement page).
Acknowledgements
Thanks to all who have contributed to this database, especially to Professor Stefano Bertolini who verified many of the mutation entries. This work is supported in
Fig. 1. The spectrum of unique mutations (excluding major rearrangements) (n=592) across the promoter and 18 exons of LDLR found in the FH mutation database.
Fig. 2. Locations of LDLR major rearrangements. All deletions and insertions are shown except for three, a 37 bp deletion in exon 4 [5], a 87 bp deletion in exon 5 [13] and a 96 bp insertion of exon 9 [14]. Insertions are marked. Sizes of the deletions and insertions are indicated above
K.E.Heath et al./Atherosclerosis154 (2001) 243 – 246 246
part by Merck Sharp Dohme, the John Pinto Founda-tion, the British Heart Foundation (Grants RG95007 and RG93008) and MEDPED, University of Utah, Salt Lake City, Utah.
References
[1] Hobbs HH, Brown MS, Goldstein JL. Molecular genetics of the LDL receptor gene in familial hypercholesterolaemia. Hum Mu-tat 1992;1:445 – 66.
[2] Francke U, Brown MS, Goldstein JL. Assignment of the human gene for the low density lipoprotein receptor to chromosome 19: synteny of a receptor, a ligand, and a genetic disease. Proc Natl Acad Sci 1984;81:2826 – 30.
[3] Sudhof TC, Goldstein HL, Brown MS, Russell DW. The LDL receptor gene: a mosaic of exons shared with different proteins. Science 1985;228:815 – 22.
[4] Wilson DJ, Gahan M, Haddad L, Heath K, Whittall RA, Williams RR, Humphries SE, Day INM. A World Wide Web site for low-density lipoprotein receptor gene mutations in famil-ial hypercholesterolaemia: sequence based, tabular and direct submission data handling. Am J Cardiol 1998;81:1509 – 11. [5] Giesel J, Holzem G, Oette K. Screening for mutations in exon 4
of the LDL receptor gene in a German population with severe hypercholesterolaemia. Hum Genet 1995;96:301 – 4.
[6] Bertolini S. FH website, 1997.
[7] Leitersdorf E, Tobin EJ, Davignon J, Hobbs HH. Common low-density lipoprotein receptor mutations in the French Cana-dian population. J Clin Invest 1990;85:1014 – 23.
[8] Lombardi P, Sijbrands EJG, van de Giessen K, Smelt AHM, Kastelein JJP, Frants RR, Havekes LM. Mutations in the low density lipoprotein receptor gene of familial hypercholestero-laemic patients detected by denaturing gradient gel electrophore-sis and direct sequencing. J Lipid Res 1995;36:860 – 7.
[9] Nissen H, Guldberg P, Hansen A, Peterson NE, Horder M. Clinically applicable mutation screening in familial hypercholes-terolaemia. Hum Mutat 1996;8:166 – 77.
[10] Ekstrom U, Abrahamson M, Sveger T, Lombardi P, Nilsson-Ehle P. An efficient screening procedure detecting six novel mutations in the LDL receptor gene in Swedish children with hypercholesterolaemia. Hum Genet 1995;96:147 – 50.
[11] Gudnason V, King-Underwood L, Seed M, Sun X-M, Soutar AK, Humphries SE. Identification of recurrent and novel muta-tions in exon 4 of the LDL receptor gene in patients with familial hypercholesterolaemia in the United Kingdom. Arterioscler Thromb 1993;13:56 – 63.
[12] Yamamoto T, Davis CG, Brown MS, Schneider WJ, Casey ML, Goldstein JL, Russell DW. The human LDL receptor: a cys-teine-rich protein with multiple Alu sequences in its mRNA. Cell 1984;39:27 – 38.
[13] Schluter G, Wick U. An 87 bp deletion in exon 5 of the LDL receptor gene in a mother and her son with familial hypercholes-terolaemia. Clin Genet 1994;43:84 – 7.
[14] Kuhrova V. FH website submission, 1999.