Members of the
Arabidopsis
14-3-3 gene family trans-complement
two types of defects in fission yeast
Takashi Kuromori
a,1, Masayuki Yamamoto
a,b,*
aDi6ision of Cell Proliferation,National Institute for Basic Biology,Okazaki,Aichi444-0867Japan
bDepartment of Biophysics and Biochemistry,Graduate School of Science,Uni6ersity of Tokyo,Hongo,Tokyo113-0033,Japan
Received 13 March 2000; received in revised form 15 June 2000; accepted 15 June 2000
Abstract
14-3-3 proteins are highly conserved among eukaryotes and perform diverse biochemical activities. We isolated five types of
Arabidopsis14-3-3 cDNAs in a screen for clones that could block ectopic meiosis driven by the pat1 mutation in fission yeast. Overexpression of fission yeastrad24, which encodes a 14-3-3 protein, also suppressedpat1. AllArabidopsisclones isolated could rescue the deformed morphology and elevated UV sensitivity of the rad24 mutant. Thus, it appears that Arabidopsis 14-3-3 proteins can generally substitute for their fission yeast counterpart in function. Expression of anArabidopsis14-3-3 clone, GF14m, was shown to be rather ubiquitous among plant organs. © 2000 Elsevier Science Ireland Ltd. All rights reserved.
Keywords:Arabidopsis thaliana;Schizosaccharomyces pombe; 14-3-3; Trans-complementation; UV sensitivity; Pat1 kinase
www.elsevier.com/locate/plantsci
1. Introduction
14-3-3 proteins, which were originally isolated as proteins abundant in mammalian brain, are widely distributed among various organisms in-cluding mammals, yeast and plants. In mammals, they fulfil diverse functions and associate with many cellular molecules [1,2]. In plants, genes encoding 14-3-3 homologues have been isolated from various species, namely maize, barley, to-bacco, spinach, rice, tomato and others [3 – 5]. Especially, much information has been accumu-lated on Arabidopsis 14-3-3 isoforms [3 – 5].
Arabidopsis appears to have at least ten 14-3-3 isoforms [4]. GF14v was the first example of the isoforms, which was isolated originally as a
com-ponent of a protein complex bound to the tran-scription-regulating DNA element named G-box [6]. Subsequently, Arabidopsis 14-3-3 isoforms have been shown to interact with diverse cellular molecules, as do mammalian 14-3-3 proteins [3 – 5,7]. Several Arabidopsis isoforms bind to the hinge 1 region of nitrate reductase and inhibit its activity [8]. A couple of isoforms can bind and activate Arabidopsis AHA2, a plasma membrane H+-ATPase [9,10], and CPK-1, a
calcium-depen-dent protein kinase isoform [11]. Another isoform, AFT1 (GF14l), binds to ascorbate peroxidase and
O-methyltransferase [12,13]. AFT1 is also able to activate transcription in the yeast two-hybrid sys-tem [14], and immunocytochemistry shows that
Arabidopsis 14-3-3 proteins are present in the nu-clei [15]. Recently, fiveArabidopsis14-3-3 isoforms were shown to have specific affinities for transcrip-tional factor IIB in vitro [16].
The budding yeast Saccharomyces cere6isiaehas two 14-3-3 proteins, encoded by BMH1 and
BMH2, which appear to be involved in growth regulation [17,18]. The fission yeast Schizosaccha -* Corresponding author. Present address: Department of
Bio-physics and Biochemistry, Graduate School of Science, University of Tokyo, Hongo, Tokyo 113-0033, Japan. Tel.:+81-3-38149620; fax:
+81-3-58022042.
E-mail address: [email protected] (M. Yamamoto).
1Present address: Plant Functional Genomics Research Group,
RIKEN Genomic Sciences Center, Tsukuba, Ibaraki 305-0074, Japan.
romyces pombe also appears to have two 14-3-3 proteins, and they have been shown to be involved in DNA damage checkpoint control during the cell cycle [19]. It has been reported that a nuclear export signal in the S. pombe14-3-3 homologue is essential for shuttling Cdc25 out of the nucleus in a phosphorylation- and CRM1-dependent manner following DNA damage [20,21].
By trans-complementation of a fission yeast mu-tation affecting the cell cycle switch from mitosis to meiosis, we isolated a group of Arabidopsis
cDNAs, each of which encoded a member of the 14-3-3 protein family. We examined whether ex-pression of theseArabidopsis14-3-3 proteins could complement the deletion mutation in S. pombe rad24, one of the two genes encoding fission yeast 14-3-3 proteins. In addition to these results, we present some features of GF14m, which have not previously been fully described.
2. Materials and methods
2.1. Yeast and Arabidopsis strains
The following fission yeast strains were used in this study: JY712 (h+ leu1 pat1-114), as the host
for the original screening; JX379 (h− ade6 leu1
ura4-D18 rad24::ura4+), as the rad24 deletion
strain; and JY333 (h− ade6-M216 leu1), as the
control rad24+ strain. Arabidopsis thaliana
(Co-lumbia ecotype) was used as the source for RNA preparation.
2.2. Transformation of fission yeast
Transformation of S. pombe cells was carried out by electroporation as previously described [22]. The nmt1 promoter on pREP vectors, al-though it is repressible by thiamine [23], was used as a constitutive promoter throughout this study, depleting thiamine from the medium.
2.3. DNA sequencing
For sequence analysis, DNA fragments cloned from Arabidopsis in pREP3 were recloned in pBluescript KS(+) and SK(+) (Stratagene). Unidirectional deletion of DNA fragments was performed with exonuclease III and S1 nuclease (Takara Shuzo), according to the method of
Henikoff [24]. Single-stranded template DNA was prepared using the helper M13KO7 bacteriophage. Nucleotide sequencing was performed by the chain termination method of Sanger et al. [25], using a DNA sequencer (Applied Biosystems 373A). The sequence data for GF14m, deposited to the DDBJ/ EMBL/GenBank databases (accession number AB011545), has been determined in both direc-tions at least once.
2.4. UV sensiti6ity
We examined UV sensitivity of JX379 (rad24) transformed by each of the Arabidopsis 14-3-3 clones or by the vector pREP3. JY333 (rad24+)
carrying the vector pREP11 was examined as a control. pREP3 and pREP11 have different multi-cloning sites but are otherwise the same. Cells of each strain, grown in minimal medium (MM) [26], were plated on SSA agar plates [27] at (1 – 9)×102
cells per plate and immediately exposed to a vari-ety of UV dose, using a UV-linker apparatus (Funakoshi FS-800). The survival rate was deter-mined by counting the number of colonies on each plate and comparing it with unexposed controls after incubation at 30°C for 5 days.
2.5. Northern blot analysis
To detect transcripts for GF14m, RNA was extracted from flower buds, flowers, stems and leaves of mature plants, using the phenol method [28]. RNA from total plant bodies grown for various periods was also subjected to the analysis. RNA blotting and subsequent hybridization were carried out according to a standard protocol [29]. A 0.25-kb Bsp1286I fragment (one Bsp1286I site derived from the cloning vector) that covered most of the GF14m3%non-coding region was used as the
probe.
3. Results
3.1. Isolation of Arabidopsis clones that inhibit ectopic meiosis in the fission yeast pat1 mutant
The pat1(ran1) mutation was originally iden-tified as an inducer of uncontrolled meiosis in S.
en-tering meiosis during vegetative growth. Pat1 ki-nase undergoes inactivation when cells are directed to enter meiosis [32,33]. Nutritional starvation is normally required for fission yeast cells to initiate meiosis, but the temperature-sensitivepat1 mutant enters the meiotic pathway even in rich medium if shifted to the restrictive temperature [30,31].
We screened for Arabidopsis cDNA clones that could repress the uncontrolled meiosis driven by the pat1ts mutation. We used Arabidopsis cDNA
libraries that could be expressed in fission yeast, as described previously [34]. More than 30Arabidop
-sis cDNA clones were isolated in this screening, and partial nucleotide sequencing of them sug-gested that the deduced gene products included five kinds of 14-3-3 proteins. They appeared to correspond toArabidopsis14-3-3 isoforms GF14x, GF14f, GF14m, GF14y, and GF14v (data not shown). The sequence data obtained for GF14m
have been deposited to the DDBJ/EMBL/Gen-Bank databases under accession number AB011545, because it is slightly different from, and somewhat longer than, the previous report (see later).
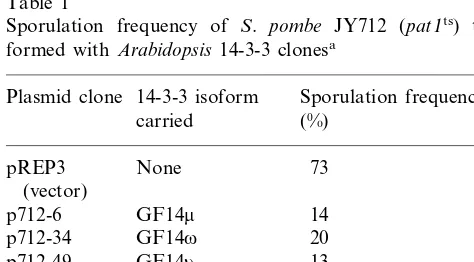
The ability of the Arabidopsis 14-3-3 clones to suppress the pat1 mutation was examined more precisely. Table 1 summarizes sporulation effi-ciency of pat1 mutant cells transformed with each of these clones. Whereas control pat1 cells carry-ing the vector pREP3 showed a high rate of ectopic meiosis and subsequent sporulation at the restrictive temperature, cells transformed with any
of the Arabidopsis14-3-3 clones were considerably inhibited from ectopic sporulation under the same conditions. All the Arabidopsis clones suppressed
pat1 nearly to the same level.
The two genes encoding 14-3-3 protein homo-logues in fission yeast, namely rad24 and rad25, function primarily in the DNA damage checkpoint [19]. Because it was unknown whether the over-production of authentic S. pombe 14-3-3 protein could suppress the pat1 mutation, we overex-pressed rad24 in the pat1 mutant. As shown in Table 1, rad24 could suppress pat1 a little more efficiently than the Arabidopsis 14-3-3 clones.
3.2. Expression of Arabidopsis 14-3-3 genes restores normal cell morphology and UV resistance to the fission yeast rad24 mutant
We examined more directly whether the Ara
-bidopsis 14-3-3 clones could perform the function of S. pombe14-3-3. It has been shown that rad24 null cells are UV sensitive, have unusual cone-shaped morphology, and enter mitosis prema-turely, which results in reduction of cell size at division [19]. We used rad24 defective cells for the analysis, since they show a stronger phenotype in UV sensitivity than rad25 defective cells [19]. We transformed the rad24 deletion mutant with Ara
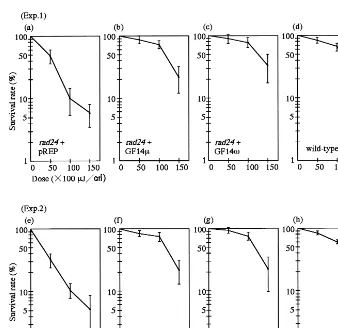
-bidopsis 14-3-3 clones. Compared with rad24 de-fective cells carrying the vector pREP3, cells carrying any of the five Arabidopsis 14-3-3 clones became longer in size. They were either indistin-guishable from or even larger than the wild type. A typical picture of the GF14m transformant is shown in Fig. 1. The 14-3-3 transformants were also as resistant to UV as the wild type, while the parental rad24 deletion strain showed obvious UV sensitivity, due to the deficiency in radiation checkpoint control (Fig. 2). These results, together with the results given in the previous section, lead us to conclude that any of the five Arabidopsis
14-3-3 proteins can fulfil, at least partially, the function of the 14-3-3 protein encoded byrad24 in fission yeast.
3.3. Characterization of GF14m
When we determined the sequences of the Ara
-bidopsis 14-3-3 clones isolated in this study, GF14m had not been reported in the literature. Therefore, we carefully sequenced this clone. Our
Table 1
Sporulation frequency of S. pombe JY712 (pat1ts)
trans-formed withArabidopsis14-3-3 clonesa
Sporulation frequency
aCells of thepat1 mutant (JY712) were transformed with
Fig. 1. Cell morphology of fission yeast strains JY333 (wild type) carrying the pREP vector (A), JX379 (rad24) carrying the pREP vector (B), and JX379 carrying p712-6 (C). Cells growing exponentially in MM liquid medium were observed under the phase-contrast microscope. Bar, 10mm.
Fig. 2. UV sensitivity of fission yeast strains JY333 (wild type) carrying the pREP vector, JX379 (rad24) carrying the pREP vector, and JX379 carrying variousArabidopsis14-3-3 clones. The dose of UV irradiation is given on the abscissa, and survival percentage is plotted on the ordinate. (a), (e) JX379 with the vector; (b) JX379 with p712-6 (GF14m); (c) JX379 with p712-34 (GF14v); (d) JY333 (wild type) with the vector; (f) JX379 with p712-49 (GF14y); (g) JX379 with p712-55 (GF14x); and (h) JX379 with p712-81 (GF14f). The measurement for each strain was repeated either ten times (a – d) or five times (e – h), and values are the means9S.D.
data (accession number AB011545) subsequently turned out to be identical to the cDNA clone registered by Ferl et al. as GF14m(accession num-ber ATU60444) except for two nucleotide
As described in Section 1, GF14x shows appar-ent organ-specific distribution [35]. Expression of GF14v is reported to be relatively low in leaves [6]. GF14f, GF14yand GF14oare also inferred to display a certain degree of organ-specific expres-sion patterns [4]. Since no data were available about expression of GF14m, we examined it by RNA blot analysis. To see tissue-specific expres-sion of GF14m, RNA was extracted from flower buds, flowers, stems and leaves of adultArabidop
-sis plants. RNA was also extracted from total plants grown for 10, 20, 30, and 40 days after seeding, to examine developmental stage-specific expression of GF14m. To exclude cross-hybridiza-tion of transcripts derived from other 14-3-3 iso-form genes, a probe was chosen that would detect part of the 3% non-coding sequence of the GF14m
transcript, which had little homology with other GF14 subtypes. Northern blot analysis revealed that this isoform is transcribed into a single mRNA species of about 1.2 kb. This transcript appeared to be abundant in all organs examined, and no obvious organ- or developmental stage-specific distribution was observed (Fig. 3).
4. Discussion
It has been previously shown that four Ara
-bidopsis 14-3-3 isoforms can complement lethality of the bmh1 bmh2 double disruptant in budding
yeast [18]. The present study indicates that Ara
-bidopsis 14-3-3 can be functional also in fission yeast, an organism distantly related to budding yeast. The five Arabidopsis 14-3-3 isoforms ana-lyzed in this study behaved similarly in trans-com-plementation of S. pombe pat1 and rad24 mutations. Although Arabidopsis 14-3-3 isoforms are thought to be somewhat diverse phylogenically [4], they all have nearly identical amino acid se-quences in the five evolutionarily conserved blocks, four of which correspond to alpha-helical regions [36]. These observations suggest that most, if not all, of 14-3-3 isoforms share common func-tional characteristics. Many of 14-3-3 associated proteins bind to 14-3-3 in a phosphorylation-de-pendent manner [37,38]. Structural analysis of complexes formed between 14-3-3 and phospho-peptides has identified amino acid residues in 14-3-3 that are important for phosphopeptide binding [39,40]. These residues are conserved evolutionar-ily from yeast to human, and the residues sup-posed to be involved in direct peptide binding are also conserved in each Arabidopsis14-3-3 isoform. It has been recently shown that a nuclear export signal exists in 14-3-3 proteins [41], and this is also the case with Arabidopsis isoforms.
We obtained Arabidopsis 14-3-3 cDNA clones as suppressors of uncontrolled meiosis driven by the pat1 ts mutation in fission yeast. The
mecha-nism for this suppression is yet unclear, but one possible explanation could be that, like mitotic arrest caused by DNA damage checkpoint control [20,21], exclusion of Cdc25 from the nucleus by 14-3-3 may arrest progression of meiosis. Cdc25 protein phosphatase activates Cdc2 kinase by dephosphorylating a tyrosine residue, which is a key step to initiate mitotic events. Cdc25 is known to be necessary for meiosis I and II in fission yeast [42,43]. The observation that S. pombecells over-producing Arabidopsis 14-3-3 isoforms are larger in cell length than the wild-type (Fig. 1) suggests that they may be delayed in mitosis because Cdc25 cannot function at its full activity.
Although it is possible that Arabidopsis 14-3-3 interacts with fission yeast Cdc25 and suppresses its activity, as already discussed, a more plausible explanation has been emerging from our recent study. Pat1 is a kinase that phosphorylates and inactivates the meiotic regulator Mei2 in fission yeast [44]. We have found that Rad24, a 14-3-3 homologue in fission yeast, physically interact with
Mei2 (M. Sato et al., unpublished results). This raises an interesting possibility that suppression of meiosis by 14-3-3 may be performed through inhi-bition of the activity of Mei2. Further analysis is in progress to examine this hypothesis.
The findings of this study apparently lead to an interesting question whether Arabidopsis 14-3-3 proteins have any intrinsic role in regulation of the progression of meiosis and/or in checkpoint con-trol over the cell cycle in plants. Given the func-tional conservation of 14-3-3 isoforms in a wide range of eukaryotes, further comparative analysis of 14-3-3 function in plants and unicellular yeast species will provide clues to this key question.
Acknowledgements
We thank Dr R. Haselkorn for critical reading of the manuscript. This work was supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Science, Sports and Cul-ture of Japan to M.Y.
References
[1] A. Aitken, 14-3-3 proteins on the MAP, Trends Biochem. Sci. 20 (1995) 95 – 97.
[2] A. Aitken, 14-3-3 and its possible role in co-ordinating multiple signalling pathway, Trends Cell Biol. 6 (1996) 341 – 347.
[3] R.J. Ferl, 14-3-3 proteins and signal transduction, Annu. Rev. Plant Physiol. Plant Mol. Biol. 47 (1996) 49 – 73. [4] K. Wu, M.F. Rooney, R.J. Ferl, TheArabidopsis14-3-3
multigene family, Plant Physiol. 114 (1997) 1421 – 1431. [5] C. Finnie, J. Borch, D.B. Collinge, 14-3-3 proteins: eukaryotic regulatory proteins with many functions, Plant Mol. Biol. 40 (1999) 545 – 554.
[6] G. Lu, A.J. DeLisle, N.C. de Vetten, R.J. Ferl, Brain proteins in plants: an Arabidopsis homolog to neuro-transmitter pathway activators is part of a DNA binding complex, Proc. Natl. Acad. Sci. USA 89 (1992) 11490 – 11494.
[7] G. Lu, N.C. de Vetten, P.C. Sehnke, T. Isobe, T. Ichimura, H. Fu, G.P. van Heusden, R.J. Ferl, A single
ArabidopsisGF14 isoform possesses biochemical charac-teristics of diverse 14-3-3 homologues, Plant Mol. Biol. 25 (1994) 659 – 667.
[8] K. Kanamaru, R. Wang, W. Su, N.M. Crawford, Ser-534 in the hinge 1 region ofArabidopsisnitrate reductase is conditionally required for binding of 14-3-3 proteins and in vitro inhibition, J. Biol. Chem. 274 (1999) 4160 – 4165.
[9] T. Jahn, A.T. Fuglsang, A. Olsson, I.M. Bruntrup, D.B. Collinge, D. Volkmann, M. Sommarin, M.G. Palmgren,
C. Larsson, The 14-3-3 protein interacts directly with the C-terminal region of the plant plasma membrane H(+ )-ATPase, Plant Cell 9 (1997) 1805 – 1814.
[10] L. Baunsgaard, A.T. Fuglsang, T. Jahn, H.A. Korthout, A.H. de Boer, M.G. Palmgren, The 14-3-3 proteins associate with the plant plasma membrane H(+ )-AT-Pase to generate a fusicoccin binding complex and a fusicoccin responsive system, Plant J. 13 (1998) 661 – 671. [11] L. Camoni, J.F. Harper, M.G. Palmgren, 14-3-3 proteins activate a plant calcium-dependent protein kinase (CDPK), FEBS Lett. 430 (1998) 381 – 384.
[12] H. Zhang, J. Wang, U. Nickel, R.D. Allen, H.M. Good-man, Cloning and expression of an Arabidopsis gene encoding a putative peroxisomal ascorbate peroxidase, Plant Mol. Biol. 34 (1997) 967 – 971.
[13] H. Zhang, J. Wang, H.M. Goodman, An Arabidopsis
gene encoding a putative 14-3-3-interacting protein, caf-feic acid/5-hydroxyferulic acid O-methyltransferase, Biochim. Biophys. Acta 1353 (1997) 199 – 202.
[14] J. Wang, H.M. Goodman, H. Zhang, An Arabidopsis
14-3-3 protein an act as a transcriptional activator in yeast, FEBS Lett. 493 (1999) 282 – 284.
[15] E.A. Bihn, A.L. Paul, S.W. Wang, G.W. Erdos, R.J. Ferl, Localization of 14-3-3 proteins in the nuclei of
Arabidopsis and maize, Plant J. 12 (1997) 1439 – 1445. [16] S. Pan, P.C. Sehnke, R.J. Ferl, W.B. Gurley, Specific
interactions with TBP and TFIIB in vitro suggest that 14-3-3 proteins may participate in the regulation of transcription when part of a DNA binding complex, Plant Cell 11 (1999) 1591 – 1602.
[17] G.P.H. van Heusden, T.J. Wenzel, E.L. Lagendijk, H.Y. de Steensma, J.A. van den Berg, Characterization of the yeast BMH1 gene encoding a putative protein ho-mologous to mammalian protein kinase II activators and protein kinase C inhibitors, FEBS Lett. 302 (1992) 145 – 150.
[18] G.P.H. van Heusden, D.J. Griffiths, J.C. Ford, T.F. Chin-A-Woeng, P.A. Schrader, A.M. Carr, H.Y. de Steensma, The 14-3-3 proteins encoded by the BMH1 and BMH2 genes are essential in the yeast Saccha
-romyces cere6isiaeand can be replaced by a plant
homo-logue, Eur. J. Biochem. 229 (1995) 45 – 53.
[19] J.C. Ford, F. Al-Khodairy, E. Fotou, K.S. Sheldrick, D.J.F. Griffiths, A.M. Carr, 14-3-3 protein homologs required for the DNA damage checkpoint in fission yeast, Science 265 (1994) 533 – 535.
[20] A. Lopez-Girona, B. Furnari, O. Mondesert, P. Russell, Nuclear localization of Cdc25 is regulated by DNA damage and a 14-3-3 protein, Nature 397 (1999) 172 – 175.
[21] Y. Zeng, H. Piwnica-Worms, DNA damage and replica-tion checkpoints in fission yeast require nuclear exclu-sion of the Cdc25 phosphatase via 14-3-3 binding, Mol. Cell. Biol. 19 (1999) 7410 – 7419.
[22] H.L. Prentice, High efficiency transformation of
Schizosaccharomyces pombe by electroporation, Nucl. Acids Res. 20 (1992) 621.
[24] S. Henikoff, Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing, Gene 28 (1984) 351 – 359.
[25] F. Sanger, S. Nicklen, A.R. Coulson, DNA sequencing with chain-terminating inhibitors, Proc. Natl. Acad. Sci. USA 74 (1977) 5463 – 5467.
[26] T. Isshiki, N. Mochizuki, T. Maeda, M. Yamamoto, Characterization of a fission yeast gene, gpa2, that en-codes a G alpha subunit involved in the monitoring of nutrition, Genes Dev. 6 (1992) 2455 – 2462.
[27] R. Egel, M. Egel-Mitani, Premeiotic DNA synthesis in fission yeast, Exp. Cell. Res. 88 (1974) 127 – 134. [28] A. Watanabe, C.A. Price, Translation of mRNAs for
subunits of chloroplast coupling factor 1 in spinach, Proc. Natl. Acad. Sci. USA 79 (1982) 6304 – 6308. [29] J. Sambrook, E.F. Fritsch, T. Maniatis, Molecular
Cloning, A Laboratory Manual, Cold Spring Harbor Laboratory Press, New York, 1989.
[30] Y. Iino, M. Yamamoto, Mutants of Schizosaccha
-romyces pombe which sporulate in the haploid state, Mol. Gen. Genet. 198 (1985) 416 – 421.
[31] P. Nurse, Mutants of the fission yeast Schizosaccha
-romyces pombewhich alter the shift between cell prolif-eration and sporulation, Mol. Gen. Genet. 198 (1985) 497 – 502.
[32] M. McLeod, D. Beach, A specific inhibitor of theran1+
protein kinase regulates entry into meiosis inSchizosac
-charomyces pombe, Nature 332 (1988) 509 – 514. [33] M. Yamamoto, The molecular control mechanisms of
meiosis in fission yeast, Trends Biochem. Sci. 21 (1997) 18 – 22.
[34] T. Kuromori, M. Yamamoto, Cloning of cDNAs from
Arabidopsis thaliana that encode putative protein phos-phatase 2C and a human Dr1-like protein by transfor-mation of a fission yeast mutant, Nucl. Acids Res. 22 (1994) 5296 – 5301.
[35] C.J. Daugherty, M.F. Rooney, P.W. Miller, R.J. Ferl, Molecular organization and tissue-specific expression of
an Arabidopsis 14-3-3 gene, Plant Cell 8 (1996) 1239 – 1248.
[36] W. Wang, D.C. Shakes, Molecular evolution of the 14-3-3 protein family, J. Mol. Evol. 43 (1996) 384 – 398. [37] Y. Furukawa, N. Ikuta, S. Omata, T. Yamauchi, T. Isobe, T. Ichimura, Demonstration of the phosphoryla-tion-dependent interaction of tryptophan hydroxylase with the 14-3-3 protein, Biochem. Biophys. Res. Com-mun. 194 (1993) 144 – 149.
[38] N.R. Michaud, J.R. Fabian, K.D. Mathes, D.K. Mor-rison, 14-3-3 is not essential for Raf-1 function: identifi-cation of Raf-1 proteins that are biologically activated in a 14-3-3- and Ras-independent manner, Mol. Cell. Biol. 15 (1995) 3390 – 3397.
[39] A.J. Muslin, J.W. Tanner, P.M. Allen, A.S. Shaw, Inter-action of 14-3-3 with signaling proteins is mediated by the recognition of phosphoserine, Cell 84 (1996) 889 – 897.
[40] M.B. Yaffe, K. Rittinger, S. Volinia, P.R. Caron, A. Aitken, H. Leffers, S.J. Gamblin, S.J. Smerdon, L.C. Cantley, The structural basis for 14-3-3: phosphopeptide binding specificity, Cell 91 (1997) 961 – 971.
[41] K. Rittinger, J. Budman, J. Xu, S. Volinia, L.C. Cantley, S.J. Smerdon, S.J. Gamblin, M.B. Yaffe, Structural analysis of 14-3-3 phosphopeptide complexes identifies a dual role for the nuclear export signal of 14-3-3 in ligand binding, Mol. Cell 4 (1999) 153 – 166.
[42] B. Grallert, M. Sipiczki, Common genes and pathways in the regulation of the mitotic and meiotic cell cycles of
Schizosaccharomyces pombe, Curr. Genet. 20 (1991) 199 – 204.
[43] Y. Iino, Y. Hiramine, M. Yamamoto, The role of cdc2 and other genes in meiosis in Schizosaccharomyces pombe, Genetics 140 (1995) 1235 – 1245.
[44] Y. Watanabe, S. Shinozaki-Yabana, Y. Chikashige, Y. Hiraoka, M. Yamamoto, Phosphorylation of RNA-binding protein controls cell cycle switch from mitotic to meiotic in fission yeast, Nature 386 (1997) 187 – 190.