DNA EXTRACTION AND CHARACTERIZATION OF
Salmonella
’s
LYTIC BACTERIOPHAGE GENOME
SALEM MOHAMED EBRAEIK
GRADUATE SCHOOL
BOGOR AGRICULTURAL UNIVERSITY
BOGOR
DECLARATION
I declare that this thesis entitled “DNA Extraction and Characterization of Salmonella’s Lytic Bacteriophage Genome” is a report of research work
carried out by me through the guidance of my academic supervisors and has not been submitted in any form for another degree at any other university. Information obtained from published or unpublished work of others and help received during laboratory and field work have been acknowledged.
Bogor, June 2013
ABSTRACT
SALEM MOHAMED EBRAIEK. 2013. DNA Extraction and Characterization of Salmonella‟s Lytic Bacteriophage Genome. Under supervision of SRI BUDIARTI, Chairman and IMAN RUSMANA, Member of Advisory Committee
Bacteriophages are infectious virus that can replicate only inside their specific bacterial host. Bacteriophage can be used as a biocontrol agent for a specific bacteria such as Salmonella and other applications. Understanding of
their molecular genetics is important due to their potential applications. The objectives of this research was to study three isolates of Salmonella‟s phages and to find out the best method to isolate and characterize genomic DNA of the phages. Three different methods i.e. PEG/NaCl Method, Scraping plaque method
and Enrichment Method were used to concentrate phages for genomic DNA extraction. Quantity and purity of extracted DNA was determine based on their optical density ratio of 260/280 nm and agarose gel electrophoresis. Genomic DNA size of the phages were determined using agarose gel electrophoresis with High range ladder as a marker. The best method to prepare bacteriophage concentrates for DNA genome extraction was enrichment method. This method had higher concentration and purity of extracted DNA. Based on DNA genome size, the bacteriophage genome of Salmonella sp were comparable with other study of Salmonella phages. Phage P15 with genome size of 39,5 kb was close to phage epsilon15. Phage P19 with genome size of 41 kb was close to phage ST160, and phage P38 with genome size of 48,5 kb was close to phage Gifsy-1.
SUMMARY
SALEM MOHAMED EBRAIEK. 2013. DNA Extraction and Characterization of Salmonella‟s Lytic Bacteriophage Genome. Under supervision of SRI BUDIARTI, Chairman and IMAN RUSMANA, Member of Advisory Committee
Bacteriophages are ubiquitous in nature and are known to proliferate wherever their bacterial hosts exist. Their virion particles can exist independently outside the host, however all phages are need their host to propagate. Several phages are highly specific to host cell surface receptors and any slight changes in structure results in little or no interaction between the phage and its host.
Salmonella‟s phage isolated from sewage water where their bacteria exist.
Three isolated of phages (phages 15, 19 and 38) were used in this research that concentrated using three types of methods. PEG/ NaCl method was firstly used to concentrate the phages based on their molecular weight to precipitate the
phage particles during centrifugation. The second method was Scraping Plaque method. In this method, overlay plaques were scraped and collected in SM buffer to get phage concentrates. Enrichment method was the last method used to concentrate the phages. Phage genome isolation was done using two methods i.e. EDTA/SDS and Proteinase K methods. The first method was done using DNase I to eliminate bacterial DNA followed by SDS/EDTA treatment to release phages DNA and phenol-chloroform-isoamyl alcohol to extract the DNA. DNA was collected by isopropanol extraction. While in Proteinase K method, bacterial DNA was eliminated by DNase I and Proteinase K/SDS was used to degrade phages capsids and ethanol for DNA precipitation.
Enrichment method results were the best while scraping and PEG/NaCl method
was appeared smear background.
Based on DNA genome size determination, the bacteriphages of Salmonella sp were comparable with other study of Salmonella phages. Phage
P15 with genome size of 39,5 kb was close to phage epsilon15. Phage P19 with genome size of 41 kb was close to phage ST160. And phage P38 with genome size of 48,5 kb was close to phage Gifsy-1.
Keywords: Salmonella, phage, DNA, Enrichment method,
© Copyright by Bogor Agricultural University 2013
All rights reserved
No part or all of this thesis may be excerpted without inclusion or mentioning the sources. Excerption only for research and education use, writing for scientific papers, reporting, critical writing or reviewing of a problem
DNA EXTRACTION AND CHARACTERIZATION OF
Salmonella
’s
LYTIC BACTERIOPHAGE GENOME
SALEM MOHAMED EBRAEIK
A thesis submitted as partial fulfillment of the requirement for the degree of Master of Science in Microbiology
GRADUATE SCHOOL
Thesis Title : DNA Extraction and Characterization of Salmonella‟s Lytic Bacteriophage Genome
Name : Salem Mohamed Ebraeik Student I.D. : G351098261
Approved
Advisory Committee
Dr. dr. Sri Budiarti Dr. Ir. Iman Rusmana, M.Si Chairman Committee Member
Agreed
Head of Study Program Dean of Graduate School
Microbiology
Prof. Dr. Anja Meryandini, M.S. Dr.Ir. Dahrul Syah, M.Sc.Agr.
ACKNOWLEDGEMENT
In the name of Allah the Most Gracious, The Most Merciful. Many individuals were responsible for the crystallization of this work, whose associations and encouragement have contributed to the accomplishment of the present report, and I would like to pay tribute to all of them.
I would like to share my deepest gratitude and sincere appreciation to my advisors, Dr. dr. Sri Budiarti and Dr. Ir. Iman Rusmana for their unwavering patience, instruction, direct guidance and valuable support during my study time. I must also thank Bogor Agricultural University for the chance to study and
fulfillment the degree of Master of Science, also to Hibah Penelitian Tim Pascasarjana (HPTP) 2010 which financed this research through Dr. dr. Sri Budiarti as chairman of team Research
My sincere appreciation goes to all lab members for assistance, willingness
to share their knowledge, and friendship.
I would also like to thank Yosi Kustian, Sari Nalurita and Nani Maryani for all the discussion time and their support during my time in Bogor. I would also like to thank Microbiology students IPB 2009, 2010 for their support and friendship during my time in IPB.
Finally I would like to thank my parents, my brother and my sisters for their support and love was by far the most important element that allowed me to continue my studies.
Bogor, June 2013
Salem Mohamed Ebraeik
AUTOBIOGRAPHY
The author was born on the 17th April 1985, Misuratah, Libya. He
completed his primary education at Al-jazirah School before he joined Basic Sciences School where he completed his high school in 2004. He later joined Misuratah University and graduated with Bachelor of Science degree in Microbiology in 2007.
TABLE OF CONTENTS
Bacterial Strains and Bacteriophage Isolates... 6
Preparation of Bacteriophage Concentrate... 6
PEG/ NaCl Method... 6
Scraping Plaque Method... 7
Enrichment Method... 7
Quantification of Bacteriophages by Plaque Assay... 8
Extraction of Bacteriophage Genomic DNA... 8
DNA Extraction Using EDTA and SDS... 8
DNA Extraction Using Proteinase K... 9
Measuring Concentration and Purity of Extracted DNA... 9
Determination of Phage Genome Size... 10
RESULTS Concentration of Bacteriophage Concentrates and Their Plaque Morphology………. 11
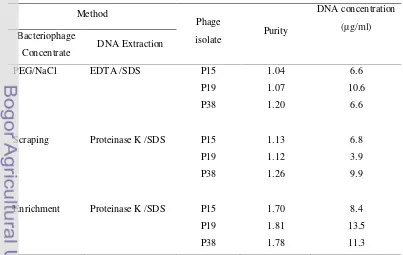
Quantity and Purity of Extracted DNA... 13
Size of Phage Genome... 14
DISCUSSION ... 16
CONCLUSION ... 18
REFERENCES ... 19
LIST OF TABLES
Page 1. Plaque morphology of phage concentrates prepared using three
different methods... 12
2. DNA concentration and purity determined using spectrophotometer of extracted DNA from Phage concentrates prepared using three different
LIST OF FIGURES
Page 1. Concentration of phage concentrates prepared using three different
methods...11
2. Plaque morphology from plaque assay of three phage concentrates prepared using (A) PEG/NaCl method, (B) Scraping plaque method,
and (C) Enrichment method... 12
3. Electrophoresis Results of phage genomic DNA extracted from phage concentrates prepared using (A) PEG/NaCl method with EDTA/SDS method, (B) Scraping method with Proteinase K/SDS method, and (C)
Enrichment method with Proteinase K/SDS…... 14
SDS Sodium dodecyl sulfate
SM buffer Solely to Tris buffer with magnesium salt ss Single stranded
SSA Salmonella Shigella Agar TAE Tris Acetate EDTA TSB Tryptone Soy Broth UV Ultraviolet
1
INTRODUCTION
Background
Bacterial pathogens are one of the most dangerous organism infecting human, animal and plant; the danger of these bacteria are related to their activity causing damage and disease to human, animal and plants (Casadevall and Pirofski 2009). Salmonella is one of bacterial pathogen that causes harmful disease (diarrhea) to human. There was estimated that 1.9-2.5 million of under 5 years old children were died from diarrhea per year in developing countries (Nguyen et al, 2006; An 2007). In developed countries: total of 155540 confirmed cases of salmonellosis were reported and 64.5% of the cases were caused by S. enteritidis in 2007 in the European Union. In United States, from 1999 salmonellesis patients caused by S. enteritidis was increased and it was more than 1400 patients in 2006 (Pan Z et al, 2009). Salmonelladerby were isolated from patients with diarrhea in Myanmar can invade Hep2 cells when tested in vitro (Budiarti et al, 1991). To fight against these bacterial pathogens, human usually use antibiotics such as
Ciprofloxacin, Norfloxacin, Nalidixic Acid and Ampicillin taken either oral or injection (Goodman et al, 1984).
2 utilized by many researchers in Eastern Europe and Russia (Huff et al. 2005).
Bacteriophage of EPEC (enteropathogenic Escherichia coli) was reported effective to control the EPEC growth (Budiarti et al. 2011). The bacteriophages of SalmonellaSp ( P15, P19, P38) isolated from domestic sewage at Darmaga Bogor
Indonesia have effectivity to lysis cells of the Salmonella sp. from diarrhea disease patient, the phage 38 was reported safe when consumed during two weeks by rat sprague dawley (Sartika et al. 2012).
The phage can be used as a biocontrol agent for a specific bacterial disease (Fischetti et al 2006). The specificity of the phage can also be used for rapid detection of specific pathogenic bacteria such as Salmonella spp and Mycobacterium tuberculosis by detecting their intracellular compounds of the lysis bacterial cells (Goodridge and Abedon 2003). The phage can also be used for tracing salmonellosis outbreaks and determining the sources of infected pathogens (Ward et al, 2005). There are other applications of bacteriophages in molecular biotechnology i.e. as delivery tools for protein and DNA vaccines, as potential gene therapy vectors and in nanotechnology techniques (Verheust et al, 2010). Therefore understanding of bacteriophages and their molecular genetics is
important due to their potential applications.
Aim of this study
The objectives of this research were to study three isolates of Salmonella‟s phages and to find out the best and easiest method to isolate and characterize
3
LITERARY REVIEW
Bacteriophages
Bacteriophages are ubiquitous in nature and are known to proliferate wherever their bacterial hosts exist (Hendrix et al. 1999). Virion particles can exist independently outside the host, however all phages are obligate intracellular parasites and need their host to propagate (Jensen et al. 1998). Several phages are highly specific to host cell surface receptors and any slight changes in structure results in little or no interaction between the phage and its host. Therefore, many phage typing schemes for the identification of bacterial species or subspecies are based on this specificity (Welkos et al. 1974).
Bacteriophages are found in almost all environments on Earth, from the depths of the ocean to hot springs, and can be isolated from almost any material that will support bacteria (Dabrowska et al. 2005). There is evidence that the diversity of bacteriophage is about an order of magnitude higher than that of bacteria (Weinbauer and Rassoulzadegan 2004) which has implications in the
classification of bacteriophage.
Bacteriophages discovered early by Frederick W. Twort and Felix
d„Herelle each one independent. However there has been considerable controversy with regards to who actually discovered the bacterial viruses first. In 1896, British bacteriologist Ernest Hankin described his observations with regards to the presence of antibacterial activity against Vibrio cholerae in the Jumna and Ganges rivers of India. He proposed that an unidentified chemical substance was responsible for the decline in the spread of cholera. A few years later, other researchers made similar observations although they did not investigate their findings further (Sulakvelidze et al.2001). Nearly 20 years after Hankin„s report, Frederick W. Twort reported on a phenomenon referred to as the „glassy
4
viruses‟ that resulted in the death of bacteria (Summers 2001). He proposed the
name „bacteriophage‟ from „bacteria‟ and „phagein‟ (Greek word for to eat or devour) therefore implying that bacteriophages „eat‟ bacteria. D„Herelle believed that a phage was an obligate parasite which is particulate, invisible, filterable, and self-reproducing in nature (Stent 1963).
Structure of Salmonella’s Bacteriophage
Capsid
The capsid is icosahedral in shape, with rare elongated variations. It appears smooth under electron microscopy and ranges in diameter from 34 to 160nm with a majority at 60nm. Capsomers (the morphological subunits) are also present (Ackermann 2003; Bradley, 1967). The family Myoviridae are generally larger than Podoviridae and Siphoviridae families (Ackermann 1998).
Tail
5 Other Structures
There is a small disk located inside the head at the site of tail attachment which is known as the connector. The connector holds the head and tail together and has functions in head assembly and DNA encapsidation. Tailed bacteriophage can also have base plates, tail spikes and tail fibres, though the number and shape of these can vary (Ackermann 1998). mechanism, where the genome is continually copied into the capsid until it is full (Streisinger et al, 1967). DNA may be concatemeric (head-to-tail repeats of a sequence) or unit length prior to packaging with concatemeric DNA formed from recombination between linear DNAs or rolling circle replication. Cleavage of concatemeric DNA for packaging can occur at; a) unique sites resulting in blunt
termini or cos ends with packaging starting and finishing at a cos site, b) pac sites (sequences recognised by terminase complex) to produce DNA molecules with limited circular permutation and terminal redundancy (excess coding DNA at the terminal end) and where packaging starts at the pac site and continues until the head is full or c) random sites to produce circularly permuted, terminally
redundant DNA (Ackermann, 1998; Ackermann 2003; Maniloff and Ackermann 1998).
6
MATERIALS AND METHODS
Time and Place of Study
This research was carried out from September 2011 to December 2012 at Biotechnology of Animal and Biomedical Laboratory, Study Center for Life Sciences and Biotechnology and Microbiology Laboratory, Biology Department, Faculty of Science and Mathematics, Bogor Agricultural University. While past of this research was carried out at Indonesian Institut of Sciences, Cibinong.
Bacterial Strains and Bacteriophage Isolates
Salmonella spp and their bacteriophages used in this study were collections of Dr. Sri Budiarti, Department of Biology Bogor Agricultural University. Three isolates of Salmonella spp were used and maintained on SSA media. Culturing Salmonella spp were on TSB or NB and incubated at 37C° for 16-24h until their concentration reached to OD600nm=1 that (1x109 CFU/ml). The bacteriophages were stored in SM buffer. The bacteriophage stocks can be
stored up to 5 months.
Preparation of Bacteriophage Concentrate
Three different types of methods were tried for preparing bacteriophage concentrates. The methods were method using Polyethilene glycol PEG/NaCl (PEG/NaCl Method), scraping of plaque on soft agar (Plaque Scraping Method), and enrichment in broth media (Enrichment Method).
PEG/ NaCl Method
7 10% and 1 M respectively. The mixture was stored at 4 C° overnight. The phage particles were centrifuged at 10,000 g for 20 min and the pellet was resuspended in 5 ml TM buffer. The remaining of PEG 8000 and cell debris was extracted from bacteriophage suspension by adding an equal volume of chloroform followed by gently vortex for 30 seconds. The organic and aqueous phases were separated by centrifugation at 3000g for 15 minutes at 4C°. Then concentration of bacteriophage particles was checked using Double Layers Agar Plaque Assay (Seyediras et al 1991; Sambrook and Russell 2001).
Scraping Plaque Method
Six petri dishes of plaque assay were prepared for each bacteriophage with suspension dilution of 10-3 and two petri dishes of each bacteriophage were scraped their overlay plaques and diluted in 3 ml of SM buffer. The suspension was mixed thoroughly using a Vortex for 30s and it was incubated at room temperature for 1-2 h to allow bacteriophage to diffuse from agar. Centrifugation was done at 4000g and 4 C° for 10 minutes and the supernatant was transferred to sterile tubes. The bacteriophage concentration was checked using Double Layers Agar Plaque Assay (Sambrook and Russell 2001).
Enrichment Method
One Petri dish of each bacteriophage isolates was prepared with low bacteriophage dilution. The plaque formation was checked after 7-9 h to affirm that the plaques were already formed then they were incubated overnight at 37 C°. Four ml of SM buffer was added on the soft top agar followed by incubation overnight at 4 C°. The SM buffer was transferred and the bacterial debris was removed by centrifugation at 4000g for 10 minutes at 4 C°. Supernatant was transferred to sterile tubes as a stock of bacteriophages.
8 of TSB was added and incubated at 37 C° for 16-24 h. The suspension was centrifuged at 4000 g and 4 C° for 10 minutes. 200 µl of supernatant was taken and incubated with 200 µl of concentrated Salmonella culture. Bacteriophage suspension was incubated at 37 C° for 20 minutes. And 2 ml of TSB media was added followed by overnight incubation. Concentration of the bacteriophage was checked by plaque assay (Sambrook and Russell 2001).
Quantification of Bacteriophages by Plaque Assay
Bacteriophage suspensions were diluted serially in NB media. And 100 μl of desired dilution of phages was incubated with 100 µl of specific strain of Salmonella at 37 C° for 20 min to allow the bacteriophage particles to attach
bacterial cells of Salmonella. Suspension was added to a tube of overlay medium and mixed throughly. The mixture was poured over underlay plate. The plates were incubated at 37C° for 18–24 h. The plaques were counted on plates. The result was accepted from plates with 30-300 plaques. The titer of the original phage preparation was determined using the following calculation (Clokie and Kropinski 2009):
Phage Concentration (pfu/ml) = Number of plaques × 10 × reciprocal of counted dilution
Extraction of Bacteriophage Genomic DNA
Two different methods for DNA isolation of phage genomes were used. The methods were DNA extraction using EDTA and SDS and DNA extraction using Proteinase K.
DNA Extraction Using EDTA and SDS
9 upper aqueous phase was taken. DNA was precipitated using isopropanol by centrifugation at 10,000g. The pellet was washed by 1 ml of ethanol 70% at room temperature and then centrifuged at 10,000g for 5 minutes. The pellet was dried and added 20 µl TE buffer. 2 µl of RNase was added and incubated for 30min at 37C°. DNA was stored at 4C° (Oakey and Owens 2000; Sambrook and Russell 2001).
DNA Extraction Using Proteinase K
Bacteriophage DNA was extracted by adding 5 µl of DNase I to 0,5 ml of bacteriophage suspension, and incubated at 37C° for 30 minutes. Proteinase K (1 mg/ml) was added to a final concentration of 50 µg/ml and SDS to a final concentration of 0.5%. The solution was mixed by vortex for a few seconds. Incubation was done at 56 C° for an hour. One volume of equilibrated phenol was added and mixed well by gently inverting. The phases of complex were separated by centrifugation at 4000g for 10 minutes at room temperature. Aqueous phage
Measuring Concentration and Purity of Extracted DNA
Concentration and purity of extracted DNA was determined using a spectrophotometer at 260/280nm. The DNA concentration was calculated from acceptable value of OD (OD 1.0 = 50 µg/ml of DNA). Spectrophotometer measurement was between 0.1–1.0. Optical density ratio of 260/280 nm were used to estimate DNA purity. Good DNA purity is at the ratio of 1.8 to 2.0 (Fong 2008).
10 1× TAE. DNA ladder was One Kb DNA ladders were used as DNA size marker.
Electrophoresis was set up at 60 volt for 50 minutes. DNA staining used 1 mg/mL ethidium bromide. DNA gels were visualized and photographed under a UV Transilluminator (Fong 2008).
Determination of Phage Genome Size
11
RESULTS
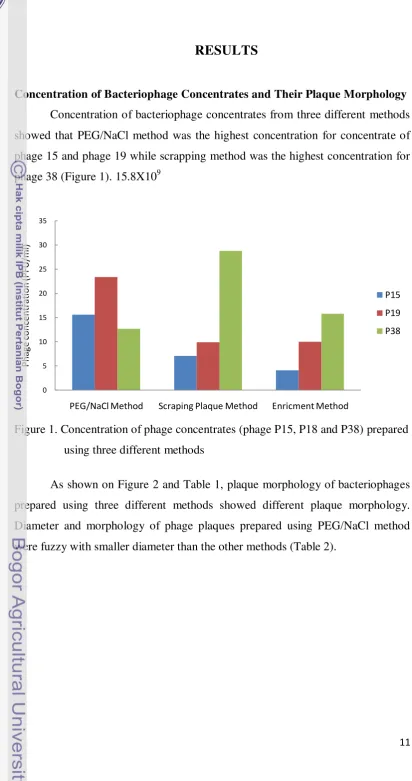
Concentration of Bacteriophage Concentrates and Their Plaque Morphology Concentration of bacteriophage concentrates from three different methods showed that PEG/NaCl method was the highest concentration for concentrate of phage 15 and phage 19 while scrapping method was the highest concentration for phage 38 (Figure 1). 15.8X109
Figure 1. Concentration of phage concentrates (phage P15, P18 and P38) prepared using three different methods
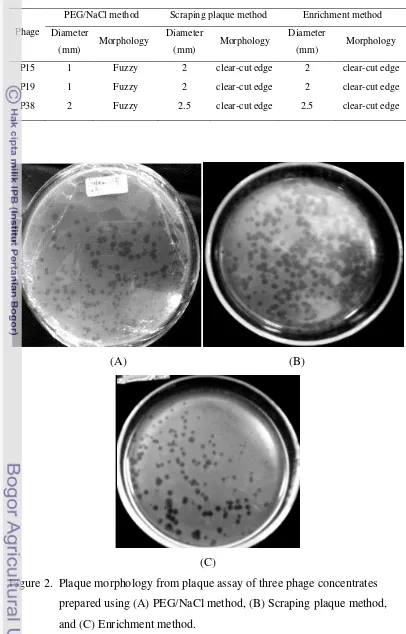
As shown on Figure 2 and Table 1, plaque morphology of bacteriophages prepared using three different methods showed different plaque morphology. Diameter and morphology of phage plaques prepared using PEG/NaCl method
were fuzzy with smaller diameter than the other methods (Table 2).
12 Table 1. Plaque morphology of phage concentrates prepared using three different
methods
Phage
PEG/NaCl method Scraping plaque method Enrichment method
Diameter
(mm) Morphology
Diameter
(mm) Morphology
Diameter
(mm) Morphology
P15 1 Fuzzy 2 clear-cut edge 2 clear-cut edge
P19 1 Fuzzy 2 clear-cut edge 2 clear-cut edge
P38 2 Fuzzy 2.5 clear-cut edge 2.5 clear-cut edge
(A) (B)
(C)
13 Quantity and Purity of Extracted DNA
Results of determining quantity and purity of extracted DNA were shown on Table 3. Optical density (OD) measurement of extracted DNA for three different methods using spectrophotometer showed that PEG/NaCl and Scraping methods were low. However, OD value ratio of 260/280nm from Enrichment method was higher indicated higher purity. While the highest DNA concentration was from phage P19 prepared using Enrichment method (Table 3).
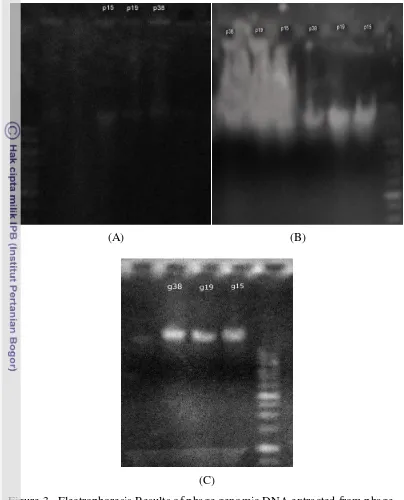
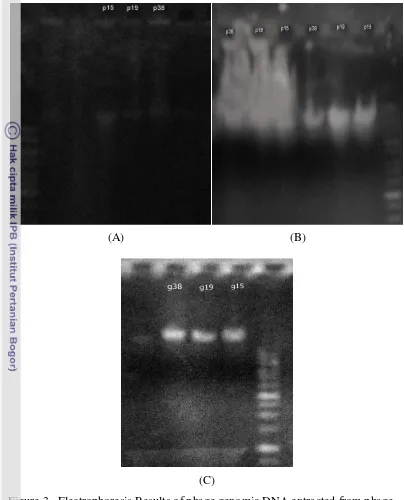
Based on agarose gel electrophoresis results (Figure 2), it was shown that effect of PEG 8000 used in precipitation of phage on agarose gel. There was appeared smear background even using chloroform to wash it, but its impact was still appeared. The same problem was appeared when using the scraping method, dusky smear was appeared because of the high concentration of agar whose had been dissolved in SM buffer. Even after washing out using phenol and chloroform three times, but its effect on electrophoresis results was still appeared. However the extracted DNA from enrichment method showed a clear DNA band indicated a good DNA purity.
14
(A) (B)
(C)
Figure 3. Electrophoresis Results of phage genomic DNA extracted from phage
concentrates prepared using (A) PEG/NaCl method with EDTA/SDS method, (B) Scraping method with Proteinase K/SDS method, and (C) Enrichment method with Proteinase K/SDS.
Size of Phage Genome
15 39,5 kb (Figure 3). While DNA genom of phage P19 was approximately 41 kb.
And DNA genom of phage P38 was approximately 48,5 kb.
Figure 3. Electrophoresis results of three phage‟s DNA genomes. M= High range DNA ladders; P38=Phage P38 genome; P19=Phage P19 genome. P15=Phage P15 genome utilizing Enrichment Method and Proteinase K /SDS.
16
DISCUSSION
In this study, three methods of isolation phages were used and compared to find that Enrichment method is the best with regard to results that obtained, and the advantages of this method are its simplicity, rapidity of isolation and reduction in the cost, In addition all chemicals used are equipments and available in the laboratories. The method is best suited for isolating DNA from a small scale.
Although using 500ml of media in PEG/NaCl method to propagate phage but concentrated phage suspension using centrifuge were low concentration comparing with large amplification occurred, that is because phage particles were often broken into head and tail components and would obviously not be viable (Oakey and Owens 2000). In contrast, the concentrates obtained through other methods contained a larger number of intact bacteriophage particles comparing with small amplification.
A few modifications were done in concentration of phages and extraction of phage DNA methods which DNase I addition was delayed until concentration of
phage particles was done in PEG/NaCl method to avoid any DNA contamination maybe happen, while RNase A was added after DNA precipitation by ethanol 70% at all of methods. Enrichment method also was modified by adding an extra time of incubation phage with bacteria, and the second incubation was added to avoid any agar transferred with first phage suspension. In addition scarping soft agar in SM buffer can be very useful which concentration of phage particulates could be increased by adding more times of soft agar from Plaque Assay to the same volume of SM buffer, that is very sample and possible but not for isolation DNA because of the difficulties to remove the high concentration of agar.
17 The chilling leads to assemble PEG molecules which precipitate by centrifugation.
This study results on the Phage DNA genome size have similarity with other published results. Phage 15 that was determined its genome size about 39,5 kb is
18
CONCLUSION
Isolation of DNA genome of Salmonella‟s phages were successfully performed for all of the three phages. The best method to prepare bacteriophage concentrates for DNA genome extraction was enrichment method. Bacteriophage concentrate from enrichment method resulted in extracted DNA of bacteriophage genome with higher purity.
19
REFERENCES
Ackermann HW. 2003. Bacteriophage observations and evolution. Res Microbiol 154: 245-51.
Ackermann HW, DuBow MS, Jarvis AW, Jones LA, Krylov VN, Maniloff J, Rocourt J, Safferman RS, Schneider J, Seldin L.1992. The species concept and its application to tailed phages. Arch Virol 124: 69-82.
Ackermann HW. (1998) Tailed bacteriophages: the Order Caudovirales. Adv Virus Res 51: 135-201.
An VT. 2007. Antibiotic resistance in Salmonella (thesis). Utrecht: Faculty of Veterinary Medicine, Utrecht University.
Bradley DE. 1967. Ultrastructure of bacteriophage and bacteriocins. Bacteriol Rev 31: 230-314.
Budiarti S. 2011. Antibiotic Resistance Escherichia coli isolated from faecal of Healthy Human. JIEAS 6 (3): 359-364.
Budiarti S, Pratiwi RH, Rusmana I. 2011. Infectivity of lytic phage to enteropathogenic Escherichia coli from Diarrheal patients in Indonesia. J US-China Med Sci 8:72-80.
Budiarti S, Hirai Y, Minami J, Katayama S, Shimizu T, Okabe A. 1991. Adherence to Hep-2 cells and replication in macrophage of Salmonella derby of human origin. Microbiol Immunol. 1991;35 (2):111-23.
Casadevall A and Pirofski L. 2009. Virulence factors and their mechanisms of action: the view from a damage–response framework. J Water Health. 07:1-18.
Casjens S. 2003. Prophages and bacterial genomics: what have we learned so far? Mol Microbiol 49: 277-300.
Chopin A, Bolotin A, Sorokin A, Ehrlich SD and Chopin M. 2001. Analysis of six prophages in Lactococcus lactis IL1403: different genetic structure of temperate and virulent phage populations. Nucleic Acids Res 29: 644-51.
Clokie MR, Kropinski AM. 2009. Bacteriophages Methods and Protocols, Volume 1: Isolation, Characterization, and Interactions. NewYork. Springer Science Business Media.
20 Duckworth DH. 1976. Who discovered bacteriophage? Bacteriol Rev 40:793-802.
Fong EG. 2008. Molecular Characterisation Of Salmonella Enterica Serovar Sofia In Australia (thesis). Melbourne. School of Applied Sciences. RMIT University.
Fischetti VA, Nelson D and Schuch R.2006. Reinventing phage therapy: are the parts greater than the sum? Nature Biotech.12:1508-1512.
Goodman LJ, Fliegelman RM, Trenholme G. M, and Kaplan RL. 1984. Comparative In Vitro Activity of Ciprofloxacin Against Campylobacter spp. and Other Bacterial Enteric Pathogens. Antimic. Ag. & Chemother. 25 (4): 504-506.
Goodridge L, Abedon ST. 2003. Bacteriophage biocontrol and bioprocessing: application of phage therapy of industry. 53:254-263.
Hendrix RW, Smith MM., Burns RN & Ford MP. 1999. Evolutionary
relationships among diverse bacteriophages and prophages: All the world„s
a phage. Proc Natl Acad Sci 96;2192-2197.
Hertveldt K, Lavigne R, Pleteneva E, Sernova N, Kurochkina L, Korchevskii R, Robben J, Mesyanzhinov V, Krylov VN and Volckaert G. 2005. Genome comparison of Pseudomonas aeruginosa large phages. J Mol Biol 354: 536-45.
Huff WE, Huff GR, Rath NC, Balog JM and Donoghue AM. 2005. Alternatives to Antibiotics: Utilization of Bacteriophage to Treat Colibacillosis and Prevent Foodborne Pathogen.Poul. Sci. 84:655–659.
Jensen EC, Schrader HS, Rieland B, Thompson TL, Lee KW, Nickerson KW & Kokjohn TA. 1998. Prevalence of broad-host range lytic bacteriophages of Sphaerotilus natans, Escherichia coli and Pseudomonas aeruginosa. Appl Environ Microbiol 64, 575-580.
Katsura I and Hendrix RW. 1984 Length determination in bacteriophage lambda tails. Cell 39: 691-8.
Kropinskia AM , Irina B, Kovalyovaa B, Billingtond CS, Patricke AN, Buttse BD, Guicharde JA, Pitchere TJ, Guthriee CC, Sydlaskee DA, Barnhille LM, Havense AK, Daye RK, Falke DR, and McConnelle MR. 2007. The
Genome of ε15, a Serotype-Converting, Group E1 Salmonella enterica-Specific Bacteriophage. Virology 369: 234–244.
21 McClelland M, Sanderson KE, Spieth J, Clifton SW, Latreille P, Courtney L, Porwollik S, Ali J, Dante M, Du F, Hou S, Layman D, Leonard S, Nguyen C, Scott K, Holmes A, Grewal N, Mulvaney E, Ryan E, Sun H, Florea L, Miller W, Stoneking T, Nhan M, Waterston R and Wilson RK. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature. 413:852-856.
Nguyen TV, Le Van P, Le Huy C, Gia KN.and Weintraub A.2006. Etiology and epidemiology of diarrhea in children in Hanoi, Vietnam. Int. J. of Infectious Dis. 10: 298-308.
Oakey HJ and Owens L. 2000. A new bacteriophage, VHML, isolated from a toxin-producing strain of Vibrio harveyi in tropical Australia. J Appl Microbiol, 89; 702-709.
Pan Z,Carter B, Garcıa JN, AbuOun M, Fookes M, Ivens A, Woodward MJ and Anjum MF. 2009. Identification of genetic and phenotypic differences associated with prevalent and non-prevalent Salmonella enteritidis phage types: analysis of variation in amino acid transport. J Microbiol, 155; 3200–3213.
Price-Carter M, Roy-Chowdhury P, Pope CE, Paine S, de Lisle GW, Collins DM, Nicol C and Carter. 2011. PE The evolution and distribution of phage ST160 within Salmonella enterica serotype Typhimurium. Source Infectious Diseases Group. 139:1262-71
.
Reddy J. 2009. A Comprehensive Method to Isolate High Quality DNA from the Cultivars of Hibiscus, J of Biotech App. 2: 0975–0943.
Sambrook J and Russell DW. 2009. Molecular Cloning: a Laboratory Manual 3rd edition. NewYork. Cold Spring Harbor Laboratory Press.
Sartika D, Budiarti S, Sudarwanto M. 2012. Phage FR38 Treatment on Sprague Dawley Rat Inferred from Blood Parameters and Organ System. HayatiJof Bioscience. Vol 19. No. 3.
Seyedirashcti S, Wood C and Akagi JM. 1991. Induction and partial purification of bacteriophages from Desulfovibrio vulgaris (Hildenborough) and Desulfovibrio desulfuricans ATCC 13541. J Gen Microbiol, 137, 1545-1549.
Stent GS. 1963. Molecular biology of bacterial viruses. San Francisco. Freeman Publishers.
22 Summers WC. 2001. Bacteriophage therapy. Annu Rev Microbiol 55, 437-451.
Sulakvelidze A, Alavidze Z & Morris JG. 2001. Bacteriophage Therapy. Antimicrob Agents Chemother 45: 649-659.
Su LH and Chiu CH. 2007. Salmonella: Clinical Importance and Evolution of Nomenclature. Chang Gung Med J, 30(3); 210-219.
Verheust C, Pauwels K, Mahillon J, Helinski DR, and Herman P. 2010. Contained Use of Bacteriophages Risk Assessment and Biosafety Recommendations. App biosafety. 1:32-44.
Vlieghea E, Phobab MF, Muyembe Tamfunb JJ and Jacobsa J. 2009. Antibiotic resistance among bacterial pathogens in Central Africa: a review of the published literature between 1955 and 2008. J. of Antimicrob Ag. 34; 295-303.
Ward MP, Brady TH, Couetil LL, Liljebjelke K, Maurer JJ, Wu CC.2005. Investigation and control of an outbreak of salmonellosis caused by multidrug-resistant Salmonella typhimurium in a population of hospitalized horses. Veterin Microbiol. 107; 233-240.
\Welkos S, Schreiber M. & Baer H. 1974. Identification of Salmonella with the 0-1 bacteriophage. Appl Microbiol 28, 618 – 622.
24 Appendix 1- Media Composition
1) Tryptone Soy Broth (TSB)
Sodium chloride 5g
Glucose 2.5g
Tryptone (Pancreatic digest of casein) 17g Soytone (Peptic digest of soybean meal) 3g Di-Potassium hydrogen phosphate 2.5g
Media components were dissolved in 1000 ml distilled water, pH was adjusted to 7.3 and broth was autoclaved.
2) Nutrient broth (NB)
Peptone 10g
Sodium chloride 5g
Beef Extract 10g
Media components were dissolved in 1000 ml distilled water, pH was adjusted to 7.3 and broth was autoclaved
25 Appendix 2- Buffers
1) TE Buffer
1M Tris-HCl (pH8.0) 10mM
EDTA 1mM
2) 50X TAE Buffer
EDTA (0.5 M) 100ml
Glacial acetic acid 57.1ml
Tris Base 242g
Buffer components were dissolved in 1000 ml distilled water
3) SM Buffer
Sodium chloride (NaCl) 5.8g
1M Tris-HCl, pH7.5 50ml
Hydrated magnesium sulfate (MgSO4-7H2O) 2g
ABSTRACT
SALEM MOHAMED EBRAIEK. 2013. DNA Extraction and Characterization of Salmonella‟s Lytic Bacteriophage Genome. Under supervision of SRI BUDIARTI, Chairman and IMAN RUSMANA, Member of Advisory Committee
Bacteriophages are infectious virus that can replicate only inside their specific bacterial host. Bacteriophage can be used as a biocontrol agent for a specific bacteria such as Salmonella and other applications. Understanding of
their molecular genetics is important due to their potential applications. The objectives of this research was to study three isolates of Salmonella‟s phages and to find out the best method to isolate and characterize genomic DNA of the phages. Three different methods i.e. PEG/NaCl Method, Scraping plaque method
and Enrichment Method were used to concentrate phages for genomic DNA extraction. Quantity and purity of extracted DNA was determine based on their optical density ratio of 260/280 nm and agarose gel electrophoresis. Genomic DNA size of the phages were determined using agarose gel electrophoresis with High range ladder as a marker. The best method to prepare bacteriophage concentrates for DNA genome extraction was enrichment method. This method had higher concentration and purity of extracted DNA. Based on DNA genome size, the bacteriophage genome of Salmonella sp were comparable with other study of Salmonella phages. Phage P15 with genome size of 39,5 kb was close to phage epsilon15. Phage P19 with genome size of 41 kb was close to phage ST160, and phage P38 with genome size of 48,5 kb was close to phage Gifsy-1.
SUMMARY
SALEM MOHAMED EBRAIEK. 2013. DNA Extraction and Characterization of Salmonella‟s Lytic Bacteriophage Genome. Under supervision of SRI BUDIARTI, Chairman and IMAN RUSMANA, Member of Advisory Committee
Bacteriophages are ubiquitous in nature and are known to proliferate wherever their bacterial hosts exist. Their virion particles can exist independently outside the host, however all phages are need their host to propagate. Several phages are highly specific to host cell surface receptors and any slight changes in structure results in little or no interaction between the phage and its host.
Salmonella‟s phage isolated from sewage water where their bacteria exist.
Three isolated of phages (phages 15, 19 and 38) were used in this research that concentrated using three types of methods. PEG/ NaCl method was firstly used to concentrate the phages based on their molecular weight to precipitate the
phage particles during centrifugation. The second method was Scraping Plaque method. In this method, overlay plaques were scraped and collected in SM buffer to get phage concentrates. Enrichment method was the last method used to concentrate the phages. Phage genome isolation was done using two methods i.e. EDTA/SDS and Proteinase K methods. The first method was done using DNase I to eliminate bacterial DNA followed by SDS/EDTA treatment to release phages DNA and phenol-chloroform-isoamyl alcohol to extract the DNA. DNA was collected by isopropanol extraction. While in Proteinase K method, bacterial DNA was eliminated by DNase I and Proteinase K/SDS was used to degrade phages capsids and ethanol for DNA precipitation.
Enrichment method results were the best while scraping and PEG/NaCl method
was appeared smear background.
Based on DNA genome size determination, the bacteriphages of Salmonella sp were comparable with other study of Salmonella phages. Phage
P15 with genome size of 39,5 kb was close to phage epsilon15. Phage P19 with genome size of 41 kb was close to phage ST160. And phage P38 with genome size of 48,5 kb was close to phage Gifsy-1.
Keywords: Salmonella, phage, DNA, Enrichment method,
24 Appendix 1- Media Composition
1) Tryptone Soy Broth (TSB)
Sodium chloride 5g
Glucose 2.5g
Tryptone (Pancreatic digest of casein) 17g Soytone (Peptic digest of soybean meal) 3g Di-Potassium hydrogen phosphate 2.5g
Media components were dissolved in 1000 ml distilled water, pH was adjusted to 7.3 and broth was autoclaved.
2) Nutrient broth (NB)
Peptone 10g
Sodium chloride 5g
Beef Extract 10g
Media components were dissolved in 1000 ml distilled water, pH was adjusted to 7.3 and broth was autoclaved
25 Appendix 2- Buffers
1) TE Buffer
1M Tris-HCl (pH8.0) 10mM
EDTA 1mM
2) 50X TAE Buffer
EDTA (0.5 M) 100ml
Glacial acetic acid 57.1ml
Tris Base 242g
Buffer components were dissolved in 1000 ml distilled water
3) SM Buffer
Sodium chloride (NaCl) 5.8g
1M Tris-HCl, pH7.5 50ml
Hydrated magnesium sulfate (MgSO4-7H2O) 2g
1
INTRODUCTION
Background
Bacterial pathogens are one of the most dangerous organism infecting human, animal and plant; the danger of these bacteria are related to their activity causing damage and disease to human, animal and plants (Casadevall and Pirofski 2009). Salmonella is one of bacterial pathogen that causes harmful disease (diarrhea) to human. There was estimated that 1.9-2.5 million of under 5 years old children were died from diarrhea per year in developing countries (Nguyen et al, 2006; An 2007). In developed countries: total of 155540 confirmed cases of salmonellosis were reported and 64.5% of the cases were caused by S. enteritidis in 2007 in the European Union. In United States, from 1999 salmonellesis patients caused by S. enteritidis was increased and it was more than 1400 patients in 2006 (Pan Z et al, 2009). Salmonelladerby were isolated from patients with diarrhea in Myanmar can invade Hep2 cells when tested in vitro (Budiarti et al, 1991). To fight against these bacterial pathogens, human usually use antibiotics such as
Ciprofloxacin, Norfloxacin, Nalidixic Acid and Ampicillin taken either oral or injection (Goodman et al, 1984).
2 utilized by many researchers in Eastern Europe and Russia (Huff et al. 2005).
Bacteriophage of EPEC (enteropathogenic Escherichia coli) was reported effective to control the EPEC growth (Budiarti et al. 2011). The bacteriophages of SalmonellaSp ( P15, P19, P38) isolated from domestic sewage at Darmaga Bogor
Indonesia have effectivity to lysis cells of the Salmonella sp. from diarrhea disease patient, the phage 38 was reported safe when consumed during two weeks by rat sprague dawley (Sartika et al. 2012).
The phage can be used as a biocontrol agent for a specific bacterial disease (Fischetti et al 2006). The specificity of the phage can also be used for rapid detection of specific pathogenic bacteria such as Salmonella spp and Mycobacterium tuberculosis by detecting their intracellular compounds of the lysis bacterial cells (Goodridge and Abedon 2003). The phage can also be used for tracing salmonellosis outbreaks and determining the sources of infected pathogens (Ward et al, 2005). There are other applications of bacteriophages in molecular biotechnology i.e. as delivery tools for protein and DNA vaccines, as potential gene therapy vectors and in nanotechnology techniques (Verheust et al, 2010). Therefore understanding of bacteriophages and their molecular genetics is
important due to their potential applications.
Aim of this study
The objectives of this research were to study three isolates of Salmonella‟s phages and to find out the best and easiest method to isolate and characterize
3
LITERARY REVIEW
Bacteriophages
Bacteriophages are ubiquitous in nature and are known to proliferate wherever their bacterial hosts exist (Hendrix et al. 1999). Virion particles can exist independently outside the host, however all phages are obligate intracellular parasites and need their host to propagate (Jensen et al. 1998). Several phages are highly specific to host cell surface receptors and any slight changes in structure results in little or no interaction between the phage and its host. Therefore, many phage typing schemes for the identification of bacterial species or subspecies are based on this specificity (Welkos et al. 1974).
Bacteriophages are found in almost all environments on Earth, from the depths of the ocean to hot springs, and can be isolated from almost any material that will support bacteria (Dabrowska et al. 2005). There is evidence that the diversity of bacteriophage is about an order of magnitude higher than that of bacteria (Weinbauer and Rassoulzadegan 2004) which has implications in the
classification of bacteriophage.
Bacteriophages discovered early by Frederick W. Twort and Felix
d„Herelle each one independent. However there has been considerable controversy with regards to who actually discovered the bacterial viruses first. In 1896, British bacteriologist Ernest Hankin described his observations with regards to the presence of antibacterial activity against Vibrio cholerae in the Jumna and Ganges rivers of India. He proposed that an unidentified chemical substance was responsible for the decline in the spread of cholera. A few years later, other researchers made similar observations although they did not investigate their findings further (Sulakvelidze et al.2001). Nearly 20 years after Hankin„s report, Frederick W. Twort reported on a phenomenon referred to as the „glassy
4
viruses‟ that resulted in the death of bacteria (Summers 2001). He proposed the
name „bacteriophage‟ from „bacteria‟ and „phagein‟ (Greek word for to eat or devour) therefore implying that bacteriophages „eat‟ bacteria. D„Herelle believed that a phage was an obligate parasite which is particulate, invisible, filterable, and self-reproducing in nature (Stent 1963).
Structure of Salmonella’s Bacteriophage
Capsid
The capsid is icosahedral in shape, with rare elongated variations. It appears smooth under electron microscopy and ranges in diameter from 34 to 160nm with a majority at 60nm. Capsomers (the morphological subunits) are also present (Ackermann 2003; Bradley, 1967). The family Myoviridae are generally larger than Podoviridae and Siphoviridae families (Ackermann 1998).
Tail
5 Other Structures
There is a small disk located inside the head at the site of tail attachment which is known as the connector. The connector holds the head and tail together and has functions in head assembly and DNA encapsidation. Tailed bacteriophage can also have base plates, tail spikes and tail fibres, though the number and shape of these can vary (Ackermann 1998). mechanism, where the genome is continually copied into the capsid until it is full (Streisinger et al, 1967). DNA may be concatemeric (head-to-tail repeats of a sequence) or unit length prior to packaging with concatemeric DNA formed from recombination between linear DNAs or rolling circle replication. Cleavage of concatemeric DNA for packaging can occur at; a) unique sites resulting in blunt
termini or cos ends with packaging starting and finishing at a cos site, b) pac sites (sequences recognised by terminase complex) to produce DNA molecules with limited circular permutation and terminal redundancy (excess coding DNA at the terminal end) and where packaging starts at the pac site and continues until the head is full or c) random sites to produce circularly permuted, terminally
redundant DNA (Ackermann, 1998; Ackermann 2003; Maniloff and Ackermann 1998).
6
MATERIALS AND METHODS
Time and Place of Study
This research was carried out from September 2011 to December 2012 at Biotechnology of Animal and Biomedical Laboratory, Study Center for Life Sciences and Biotechnology and Microbiology Laboratory, Biology Department, Faculty of Science and Mathematics, Bogor Agricultural University. While past of this research was carried out at Indonesian Institut of Sciences, Cibinong.
Bacterial Strains and Bacteriophage Isolates
Salmonella spp and their bacteriophages used in this study were collections of Dr. Sri Budiarti, Department of Biology Bogor Agricultural University. Three isolates of Salmonella spp were used and maintained on SSA media. Culturing Salmonella spp were on TSB or NB and incubated at 37C° for 16-24h until their concentration reached to OD600nm=1 that (1x109 CFU/ml). The bacteriophages were stored in SM buffer. The bacteriophage stocks can be
stored up to 5 months.
Preparation of Bacteriophage Concentrate
Three different types of methods were tried for preparing bacteriophage concentrates. The methods were method using Polyethilene glycol PEG/NaCl (PEG/NaCl Method), scraping of plaque on soft agar (Plaque Scraping Method), and enrichment in broth media (Enrichment Method).
PEG/ NaCl Method
7 10% and 1 M respectively. The mixture was stored at 4 C° overnight. The phage particles were centrifuged at 10,000 g for 20 min and the pellet was resuspended in 5 ml TM buffer. The remaining of PEG 8000 and cell debris was extracted from bacteriophage suspension by adding an equal volume of chloroform followed by gently vortex for 30 seconds. The organic and aqueous phases were separated by centrifugation at 3000g for 15 minutes at 4C°. Then concentration of bacteriophage particles was checked using Double Layers Agar Plaque Assay (Seyediras et al 1991; Sambrook and Russell 2001).
Scraping Plaque Method
Six petri dishes of plaque assay were prepared for each bacteriophage with suspension dilution of 10-3 and two petri dishes of each bacteriophage were scraped their overlay plaques and diluted in 3 ml of SM buffer. The suspension was mixed thoroughly using a Vortex for 30s and it was incubated at room temperature for 1-2 h to allow bacteriophage to diffuse from agar. Centrifugation was done at 4000g and 4 C° for 10 minutes and the supernatant was transferred to sterile tubes. The bacteriophage concentration was checked using Double Layers Agar Plaque Assay (Sambrook and Russell 2001).
Enrichment Method
One Petri dish of each bacteriophage isolates was prepared with low bacteriophage dilution. The plaque formation was checked after 7-9 h to affirm that the plaques were already formed then they were incubated overnight at 37 C°. Four ml of SM buffer was added on the soft top agar followed by incubation overnight at 4 C°. The SM buffer was transferred and the bacterial debris was removed by centrifugation at 4000g for 10 minutes at 4 C°. Supernatant was transferred to sterile tubes as a stock of bacteriophages.
8 of TSB was added and incubated at 37 C° for 16-24 h. The suspension was centrifuged at 4000 g and 4 C° for 10 minutes. 200 µl of supernatant was taken and incubated with 200 µl of concentrated Salmonella culture. Bacteriophage suspension was incubated at 37 C° for 20 minutes. And 2 ml of TSB media was added followed by overnight incubation. Concentration of the bacteriophage was checked by plaque assay (Sambrook and Russell 2001).
Quantification of Bacteriophages by Plaque Assay
Bacteriophage suspensions were diluted serially in NB media. And 100 μl of desired dilution of phages was incubated with 100 µl of specific strain of Salmonella at 37 C° for 20 min to allow the bacteriophage particles to attach
bacterial cells of Salmonella. Suspension was added to a tube of overlay medium and mixed throughly. The mixture was poured over underlay plate. The plates were incubated at 37C° for 18–24 h. The plaques were counted on plates. The result was accepted from plates with 30-300 plaques. The titer of the original phage preparation was determined using the following calculation (Clokie and Kropinski 2009):
Phage Concentration (pfu/ml) = Number of plaques × 10 × reciprocal of counted dilution
Extraction of Bacteriophage Genomic DNA
Two different methods for DNA isolation of phage genomes were used. The methods were DNA extraction using EDTA and SDS and DNA extraction using Proteinase K.
DNA Extraction Using EDTA and SDS
9 upper aqueous phase was taken. DNA was precipitated using isopropanol by centrifugation at 10,000g. The pellet was washed by 1 ml of ethanol 70% at room temperature and then centrifuged at 10,000g for 5 minutes. The pellet was dried and added 20 µl TE buffer. 2 µl of RNase was added and incubated for 30min at 37C°. DNA was stored at 4C° (Oakey and Owens 2000; Sambrook and Russell 2001).
DNA Extraction Using Proteinase K
Bacteriophage DNA was extracted by adding 5 µl of DNase I to 0,5 ml of bacteriophage suspension, and incubated at 37C° for 30 minutes. Proteinase K (1 mg/ml) was added to a final concentration of 50 µg/ml and SDS to a final concentration of 0.5%. The solution was mixed by vortex for a few seconds. Incubation was done at 56 C° for an hour. One volume of equilibrated phenol was added and mixed well by gently inverting. The phases of complex were separated by centrifugation at 4000g for 10 minutes at room temperature. Aqueous phage
Measuring Concentration and Purity of Extracted DNA
Concentration and purity of extracted DNA was determined using a spectrophotometer at 260/280nm. The DNA concentration was calculated from acceptable value of OD (OD 1.0 = 50 µg/ml of DNA). Spectrophotometer measurement was between 0.1–1.0. Optical density ratio of 260/280 nm were used to estimate DNA purity. Good DNA purity is at the ratio of 1.8 to 2.0 (Fong 2008).
10 1× TAE. DNA ladder was One Kb DNA ladders were used as DNA size marker.
Electrophoresis was set up at 60 volt for 50 minutes. DNA staining used 1 mg/mL ethidium bromide. DNA gels were visualized and photographed under a UV Transilluminator (Fong 2008).
Determination of Phage Genome Size
11
RESULTS
Concentration of Bacteriophage Concentrates and Their Plaque Morphology Concentration of bacteriophage concentrates from three different methods showed that PEG/NaCl method was the highest concentration for concentrate of phage 15 and phage 19 while scrapping method was the highest concentration for phage 38 (Figure 1). 15.8X109
Figure 1. Concentration of phage concentrates (phage P15, P18 and P38) prepared using three different methods
As shown on Figure 2 and Table 1, plaque morphology of bacteriophages prepared using three different methods showed different plaque morphology. Diameter and morphology of phage plaques prepared using PEG/NaCl method
were fuzzy with smaller diameter than the other methods (Table 2).
12 Table 1. Plaque morphology of phage concentrates prepared using three different
methods
Phage
PEG/NaCl method Scraping plaque method Enrichment method
Diameter
(mm) Morphology
Diameter
(mm) Morphology
Diameter
(mm) Morphology
P15 1 Fuzzy 2 clear-cut edge 2 clear-cut edge
P19 1 Fuzzy 2 clear-cut edge 2 clear-cut edge
P38 2 Fuzzy 2.5 clear-cut edge 2.5 clear-cut edge
(A) (B)
(C)
13 Quantity and Purity of Extracted DNA
Results of determining quantity and purity of extracted DNA were shown on Table 3. Optical density (OD) measurement of extracted DNA for three different methods using spectrophotometer showed that PEG/NaCl and Scraping methods were low. However, OD value ratio of 260/280nm from Enrichment method was higher indicated higher purity. While the highest DNA concentration was from phage P19 prepared using Enrichment method (Table 3).
Based on agarose gel electrophoresis results (Figure 2), it was shown that effect of PEG 8000 used in precipitation of phage on agarose gel. There was appeared smear background even using chloroform to wash it, but its impact was still appeared. The same problem was appeared when using the scraping method, dusky smear was appeared because of the high concentration of agar whose had been dissolved in SM buffer. Even after washing out using phenol and chloroform three times, but its effect on electrophoresis results was still appeared. However the extracted DNA from enrichment method showed a clear DNA band indicated a good DNA purity.
14
(A) (B)
(C)
Figure 3. Electrophoresis Results of phage genomic DNA extracted from phage
concentrates prepared using (A) PEG/NaCl method with EDTA/SDS method, (B) Scraping method with Proteinase K/SDS method, and (C) Enrichment method with Proteinase K/SDS.
Size of Phage Genome
15 39,5 kb (Figure 3). While DNA genom of phage P19 was approximately 41 kb.
And DNA genom of phage P38 was approximately 48,5 kb.
Figure 3. Electrophoresis results of three phage‟s DNA genomes. M= High range DNA ladders; P38=Phage P38 genome; P19=Phage P19 genome. P15=Phage P15 genome utilizing Enrichment Method and Proteinase K /SDS.
16
DISCUSSION
In this study, three methods of isolation phages were used and compared to find that Enrichment method is the best with regard to results that obtained, and the advantages of this method are its simplicity, rapidity of isolation and reduction in the cost, In addition all chemicals used are equipments and available in the laboratories. The method is best suited for isolating DNA from a small scale.
Although using 500ml of media in PEG/NaCl method to propagate phage but concentrated phage suspension using centrifuge were low concentration comparing with large amplification occurred, that is because phage particles were often broken into head and tail components and would obviously not be viable (Oakey and Owens 2000). In contrast, the concentrates obtained through other methods contained a larger number of intact bacteriophage particles comparing with small amplification.
A few modifications were done in concentration of phages and extraction of phage DNA methods which DNase I addition was delayed until concentration of
phage particles was done in PEG/NaCl method to avoid any DNA contamination maybe happen, while RNase A was added after DNA precipitation by ethanol 70% at all of methods. Enrichment method also was modified by adding an extra time of incubation phage with bacteria, and the second incubation was added to avoid any agar transferred with first phage suspension. In addition scarping soft agar in SM buffer can be very useful which concentration of phage particulates could be increased by adding more times of soft agar from Plaque Assay to the same volume of SM buffer, that is very sample and possible but not for isolation DNA because of the difficulties to remove the high concentration of agar.
17 The chilling leads to assemble PEG molecules which precipitate by centrifugation.
This study results on the Phage DNA genome size have similarity with other published results. Phage 15 that was determined its genome size about 39,5 kb is
18
CONCLUSION
Isolation of DNA genome of Salmonella‟s phages were successfully performed for all of the three phages. The best method to prepare bacteriophage concentrates for DNA genome extraction was enrichment method. Bacteriophage concentrate from enrichment method resulted in extracted DNA of bacteriophage genome with higher purity.
DNA EXTRACTION AND CHARACTERIZATION OF
Salmonella
’s
LYTIC BACTERIOPHAGE GENOME
SALEM MOHAMED EBRAEIK
GRADUATE SCHOOL
BOGOR AGRICULTURAL UNIVERSITY
BOGOR
19
REFERENCES
Ackermann HW. 2003. Bacteriophage observations and evolution. Res Microbiol 154: 245-51.
Ackermann HW, DuBow MS, Jarvis AW, Jones LA, Krylov VN, Maniloff J, Rocourt J, Safferman RS, Schneider J, Seldin L.1992. The species concept and its application to tailed phages. Arch Virol 124: 69-82.
Ackermann HW. (1998) Tailed bacteriophages: the Order Caudovirales. Adv Virus Res 51: 135-201.
An VT. 2007. Antibiotic resistance in Salmonella (thesis). Utrecht: Faculty of Veterinary Medicine, Utrecht University.
Bradley DE. 1967. Ultrastructure of bacteriophage and bacteriocins. Bacteriol Rev 31: 230-314.
Budiarti S. 2011. Antibiotic Resistance Escherichia coli isolated from faecal of Healthy Human. JIEAS 6 (3): 359-364.
Budiarti S, Pratiwi RH, Rusmana I. 2011. Infectivity of lytic phage to enteropathogenic Escherichia coli from Diarrheal patients in Indonesia. J US-China Med Sci 8:72-80.
Budiarti S, Hirai Y, Minami J, Katayama S, Shimizu T, Okabe A. 1991. Adherence to Hep-2 cells and replication in macrophage of Salmonella derby of human origin. Microbiol Immunol. 1991;35 (2):111-23.
Casadevall A and Pirofski L. 2009. Virulence factors and their mechanisms of action: the view from a damage–response framework. J Water Health. 07:1-18.
Casjens S. 2003. Prophages and bacterial genomics: what have we learned so far? Mol Microbiol 49: 277-300.
Chopin A, Bolotin A, Sorokin A, Ehrlich SD and Chopin M. 2001. Analysis of six prophages in Lactococcus lactis IL1403: different genetic structure of temperate and virulent phage populations. Nucleic Acids Res 29: 644-51.
Clokie MR, Kropinski AM. 2009. Bacteriophages Methods and Protocols, Volume 1: Isolation, Characterization, and Interactions. NewYork. Springer Science Business Media.
20 Duckworth DH. 1976. Who discovered bacteriophage? Bacteriol Rev 40:793-802.
Fong EG. 2008. Molecular Characterisation Of Salmonella Enterica Serovar Sofia In Australia (thesis). Melbourne. School of Applied Sciences. RMIT University.
Fischetti VA, Nelson D and Schuch R.2006. Reinventing phage therapy: are the parts greater than the sum? Nature Biotech.12:1508-1512.
Goodman LJ, Fliegelman RM, Trenholme G. M, and Kaplan RL. 1984. Comparative In Vitro Activity of Ciprofloxacin Against Campylobacter spp. and Other Bacterial Enteric Pathogens. Antimic. Ag. & Chemother. 25 (4): 504-506.
Goodridge L, Abedon ST. 2003. Bacteriophage biocontrol and bioprocessing: application of phage therapy of industry. 53:254-263.
Hendrix RW, Smith MM., Burns RN & Ford MP. 1999. Evolutionary
relationships among diverse bacteriophages and prophages: All the world„s
a phage. Proc Natl Acad Sci 96;2192-2197.
Hertveldt K, Lavigne R, Pleteneva E, Sernova N, Kurochkina L, Korchevskii R, Robben J, Mesyanzhinov V, Krylov VN and Volckaert G. 2005. Genome comparison of Pseudomonas aeruginosa large phages. J Mol Biol 354: 536-45.
Huff WE, Huff GR, Rath NC, Balog JM and Donoghue AM. 2005. Alternatives to Antibiotics: Utilization of Bacteriophage to Treat Colibacillosis and Prevent Foodborne Pathogen.Poul. Sci. 84:655–659.
Jensen EC, Schrader HS, Rieland B, Thompson TL, Lee KW, Nickerson KW & Kokjohn TA. 1998. Prevalence of broad-host range lytic bacteriophages of Sphaerotilus natans, Escherichia coli and Pseudomonas aeruginosa. Appl Environ Microbiol 64, 575-580.
Katsura I and Hendrix RW. 1984 Length determination in bacteriophage lambda tails. Cell 39: 691-8.
Kropinskia AM , Irina B, Kovalyovaa B, Billingtond CS, Patricke AN, Buttse BD, Guicharde JA, Pitchere TJ, Guthriee CC, Sydlaskee DA, Barnhille LM, Havense AK, Daye RK, Falke DR, and McConnelle MR. 2007. The
Genome of ε15, a Serotype-Converting, Group E1 Salmonella enterica-Specific Bacteriophage. Virology 369: 234–244.