www.elsevier.nlrlocateraqua-online
DNA cloning and characterization of an allograft
inflammatory factor-1 homologue in red sea bream
ž
Chrysophrys major
/
Masato Miyata
), Kunio Iinuma, Teruo Miyazaki
Laboratory of Aquaculture Management, Faculty of Bioresources, Fish Pathology, Mie UniÕersity,Tsu, Mie 514-8507, Japan
Received 6 January 2000; received in revised form 11 August 2000; accepted 17 September 2000
Abstract
Ž .
We isolated allograft inflammatory factor-1 AIF-1 homologue cDNA from
lipopolysaccha-Ž .
ride LPS -stimulated red sea bream leukocytes. We then determined that this cDNA encodes 444 bp in length and has an open reading frame that encodes 142 amino acids. The deduced amino
Ž .
acid sequence was homologous to the AIF-1s in carp and mammals )65.5% sequence identity . The nucleotide sequence of the protein-coding region in the AIF-1 gene was determined to have six exons and five introns. The exon–intron structure resembles those of the mouse and the human. Genomic Southern blot analysis revealed the presence of a single AIF-1 gene per haploid in red sea bream. A time course evaluation indicated that the expression level of AIF-1 gene in LPS-stimulated leukocytes was static in the first hour, but gradually increased from 3 to 24 h. The similarities between AIF-1s in mammals and red sea bream suggest that the AIF-1 in red sea bream may have a similar function in activated leukocytes as AIF-1s do in mammals.q2001 Elsevier Science B.V. All rights reserved.
Keywords: Lipopolysaccharide; AIF-1; Chrysophrys major
1. Introduction
Infectious diseases in fish cause significant losses in the commercial aquaculture industry. Investigations into the immunological mechanisms of fish are required for the
)Corresponding author.
Ž .
E-mail address: [email protected] M. Miyata .
0044-8486r01r$ - see front matterq2001 Elsevier Science B.V. All rights reserved.
Ž .
establishment of new methods for the prevention of fish diseases. Leukocytes that can be activated by various mediators are an important link in these self-defense mechanisms. Since we were interested in the effectors of leukocyte activation, we stimulated the
Ž . Ž .
leukocytes in red sea bream Chrysophrys major with lipopolysaccharide LPS . Total RNA was subjected to cDNA cloning, and a homology search on database revealed that the mRNAs from the stimulated leukocyte include several copies of a sequence that is
Ž .
homologous to the allograft inflammatory factor-1 AIF-1 .
AIF-1 has been recognized as a Ca2q-binding peptide expressed predominantly by
activated monocytes. It was originally identified in rat cardiac allografts associated with
Ž .
chronic cardiac rejection Autieri et al., 1995 . Rat AIF-1 has been described as a Ž
molecule that is expressed in macrophages, and is responsible to an IFN-g Utans et al., .
1995 . In addition, rat AIF-1 has been found in monocytes located in inflammatory
Ž .
lesions resulting from autoimmune diseases Schluesener et al., 1999 . The AIF-1s Ž
encode the EF-hand motief that plays an essential role in calcium binding Negrutskii
. Ž
and El’skaya, 1998 . The deduced amino acid sequences of AIF-1s in human Autieri,
. Ž . Ž . Ž
1996 , pig Chen et al., 1997 , mouse Watano and Kimura, 1998 and rat Utans et al.,
. X
1995 are highly conserved in not only the EF-hand region but also in both the N - and CX-ends.
In this study, we investigated the full-length sequence of AIF-1 homologous cDNA in red sea bream leukocytes. We performed Southern blot hybridization of red sea bream genomic DNA to estimate the copy number of AIF-1 genes per haploid. From red sea
Ž
bream genomic DNA, we isolated a coding region of an AIF-1 gene from initiation to .
termination codons and then we tried to identify and characterize its exon–intron structure. Also, the in vitro time course of AIF-1 gene expression was analyzed in LPS-stimulated red sea bream leukocytes.
2. Materials and methods
2.1. Isolation of leukocytes, LPS treatment, and RNA isolation
Ž . Ž .
An adult male red sea bream C. major 40 cm long was exsanguinated using a
Ž .
heperinized syringe NOVO, Denmark . Approximately 40 ml of whole blood was centrifuged at 1000=g for 5 min at room temperature. The resulting plasma was then
transferred into a new tube, incubated at 558C for 30 min, and sterilized with a 0.22-mm filter. The resulting blood cells were suspended in PBSŽy. and poured onto Ficoll 400 ŽSigma, USA . After centrifugation at 1000. =g for 5 min at room temperature, the
leukocytes remaining on the surface of the Ficoll 400 were harvested and suspended in
Ž .
RPMI 1640 media without serum Nissui, Japan . The cells were centrifuged at
1000=g for 5 min at room temperature, and then resuspended in RPMI 1640 media.
After two washings, the cells were suspended in 20 ml of fresh RPMI 1640 media
Ž .
2 Ž .
transferred into a 25-cm flask Corning, USA , incubated at 208C for 24 h, and then
Ž .
stimulated with 1 mgrml LPS Escherichia coli, 026:B6rGibco—final concentration
Ž .
for 20 h, when morphological change became rough was observed on cell surfaces.
Ž .
Total RNA was harvested with ISOGEN Nippon Gene, Japan following a standard
Ž .
technique developed by Chomczynski and Sacchi 1987 .
2.2. cDNA library
A cDNA library of LPS-stimulated leukocytes was established with TimeSaverw
Ž .
cDNA synthesis kit Pharmacia, Sweden . Total RNA was reverse-transcribed with
Ž .
oligo-dT 12–16 primer. The synthesized cDNA was introduced into l-ZapII cloning
Ž .
vector Stratagene, USA . The vector-insert was packaged with Gigapack III plus
Ž . w
Ž .
Stratagene , and transformed into bacteria using the ExAssist system Stratagene .
2.3. Sequencing and characterization
Plasmids of randomly chosen cDNA clones were isolated using an alkaline lysis
Ž .
method Sambrook et al., 1989 . The sequencing reaction for cDNA clones was performed using a dye terminator cycle sequencing kit for a DNA sequencer ABI
Ž .
Prisme 377 Perkin Elmer, USA . The sequence data was subjected to a homology
Ž .
search using the GeneBank database. http:rrwww.blast.genome.ad.jpr.
2.4. Genomic DNA isolation
Ž . Ž
Liver 100 mg was homogenized with 2 ml of 1= proteinase K buffer 500 mM
Ž . .
Tris–HCl, 100 mM EDTA, 20% wrv SDS, 100 mgrml Proteinase K . After
Ž .
incubation at 558C for 1 h, an equal volume of Tris–saturated phenol pH 8.0 was
Ž .
added and mixed by slow inversion 1 cycle every 2 s over a period of 30 min. After centrifugation at 20,000=g for 5 min at 208C, the supernatant was transferred into a new tube containing 200ml of 3 M sodium acetate and 5 ml of 99% ethanol. Genomic DNA was hooked up with aAJB-shaped glass rod. The glass rod was placed into a new
Ž .
tube containing 100 ml of TE buffer 10 mM Tris–HCl, 1 mM EDTA, pH 8.0 to dissolve the adhered genomic DNA.
2.5. Amplification of AIF-1 gene coding region
Two primers, AIF-1 initiate: 5X-GGGATCCATGGACAGCACAGCTCAAGGA-3X
X X
Ž
and AIF-1 terminate: 5 -GGGATCCTCAGGGCAGGTCTGAAAAGGT-3 underlines .
indicate BamHI site , which include the initiation and termination codons, respectively, were synthesized. PCR was carried out using 100 ng of template DNA, 0.2 mM of each dNTP, 16 pmol of each primer, 5ml of 10=reaction buffer, and 1.25 units of LA-Taq
Ž .
anneal, and 10 min at 728C for extension. The samples were post-heated at 728C for 10 min. The obtained DNA fragments were completely digested with BamHI, and con-nected with plasmid vector pUC118. Recombinant plasmid was transferred into E. coli
Ž .
XL1Blue cell Stratagene . The resulting transformants were cultured separately, and the amplified recombinant plasmids were recovered using an alkaline lysis method ŽSambrook et al., 1989 . Reaction for genomic clones were performed with a Thermo.
Ž
sequenase, fluorescent-labeled, primer cycle sequencing kit Amersham life science,
. Ž .
USA for a DNA sequencer DNA 4000L Li-Cor, USA .
2.6. Southern blot analysis of red sea bream genomic DNA
Ž .
Genomic DNA 20 mg isolated from red sea bream was digested with EcoRI,
BamHI and Pst I, passed through 0.8% agarose gel, transferred to a nitrocellulose
Ž .
membrane, and subjected to capillary blotting using immobilon NC Pure Millipore . A probe was prepared using PCR-amplified AIF-1 open reading frames from red sea
Ž
bream genomic DNA by primersAAIF-1 initiateB andAAIF-1 terminateB; see Section
. w 32 x Ž .
2.5 and labeled with a- P -dATP, using a random primer labeling kit Takara . Pre-hybridization, hybridization with 32P-labeled probe, and washing were carried out
Ž .
using the method described by Sambrook et al. 1989 . The membrane was then exposed
Ž .
to X-ray film Kodak aty808C using an intensifying screen.
2.7. Time course of AIF-1 expression in LPS-stimulated leukocytes
Ž 6 .
Leukocytes 5=10 cells in 2 ml of RPMI 1640 media were placed into chambers of a 12-well plate. Media with LPS was added to chambers to a final concentration of 0.1 mgrml LPS. The cells were kept at 258C for 24 h in the absence of LPS stimulation and for 24, 12, 6, 3, 1 h, 30, 15, and 5 min in the presence of LPS stimulation. The cells
Ž were harvested at the same time. The total RNA was isolated with ISOGEN Nippon
. Gene .
The isolated RNAs were reverse-transcribed, and PCRs were performed using primers forb-actin and AIF-1. 2.5mg of total RNA was mixed with 1ml of 0.5 mgrml
Ž . Ž .
oligo dT 12–16 Pharmacia . The 10-ml mixture was heated at 708C for 10 min, and then chilled in ice immediately afterward. Reverse transcription was performed in 20ml
Ž .
of total volume including 10 ml of total RNA with oligo dT 12–16 , 2 ml of 0.1 M DTT, 1ml of 10 mM dNTPs, 2 ml of DEPC-treated distilled water, 4 ml of 5= first
Ž .
strand buffer, and 200 units of reverse transcriptase, Superscript II Gibco . The mixture was incubated at 378C for 30 min, and terminated with 30ml of 10 mM EDTA. PCRs were performed in a 10-ml reaction volume. The reagents were premixed to a total volume of 100ml, and then divided into 10 tubes; nine tubes for reaction and one tube for excess. For each 10-ml reaction mixture, 0.2ml of cDNA template, 0.1 ml of each 100 pmol primers, 0.2 ml of 25 mM dNTPs, 1ml of 10= buffer, and 0.05ml of Taq
Ž .
at 728C for extension. The samples were post-heated at 728C for 10 min. The primers for
X X
Ž . X
the b-actin probe were 5 -TTGCTGATCCACATCTGCTG-3 BACrFWD and 5
-X Ž . Ž .
ACTACCTCATGAAGATCCTG-3 B-ACrRV will produce 736 bp . The primers for the AIF-1 probe were derived from the determined cDNA: 5X
CGCTCAGAAATG-X
Ž . X X
Ž GACAGCACAGC-3 AIF-FWD and 5 -CTGCTGGGATGGAGGGCGTCTT-3
AIF-. Ž .
RV will produce 476 bp .
2.8. Calibration assay
Reverse-transcribed cDNA from leukocytes without stimulation was used for calibra-tion. The cDNA was serially diluted with sterilized water to concentrations of 120, 60, 30, and 15 ngrml. Diluted cDNA, at 2 ml each, were added to each appropriate PCR
Ž . Ž .
mixture withb-actin primers Fig. 1 or with AIF-1 primers data not shown . A total of 22, 24, 26, and 28 thermal cycles were performed for each dilution step. The results of the PCR amplifications were visualized by electrophoresis using a 2.0% agarose gel in
Ž .
half-strength TBE 45 mM Tris–borate, 2 mM EDTA with serially diluted molecular Ž
weight markers 100, 50 and 25 ng of HindIII-digested l-DNA; density values of .
560-bp bands were analyzed . The gel was stained with 1mgrml of ethidium bromide for 15 min. Electrophoregrams were photographed using a digital camera DC120 ŽKodak , and the density value of each band was estimated and analyzed with software,.
Ž .
NIH image and Excel see Fig. 1 .
Fig. 1. Calibration assay. Reverse-transcribed cDNA was serially diluted, and 22, 24, 26, and 28 thermal cycles were performed for each dilution step. The results of the PCR amplifications were visualized by
Ž .
3. Results
3.1. Sequence of AIF-1 cDNA and AIF-1 gene open reading frames
A total of 150 randomly chosen clones were sequenced with forward primer. Aside from the numerous unknown sequences that could not be identified in the homology search, the most frequent gene encountered was 12 clones of cDNA with homology to mammalian AIF-1s.
The AIF-1 homologous cDNA was 444 bp in length with an open reading frame that
Ž .
encodes 147 amino acids Fig. 2 . All of the cDNA clones encoded exactly the same sequence. The amplified PCR product from genomic DNA was single banded. The sequence of ORFs in the genomic DNA was exactly the same as the cDNA encoding sequence. The red sea bream AIF-1 gene included six exons and five introns. The ORFs were 19, 62, 67, 42, 166, and 88 bp in length from ORF1 to ORF6, respectively. All the
Ž .
intervening sequences IVS follow the GT-AG rule, and there were 93, 150, 90, 160,
Ž .
and 408 bp, from IVS1 to IVS6, respectively Fig. 3 .
Ž .
The predicted amino acid sequence of red sea bream AIF-1 DDBJrAB019540 was
Ž . Ž
homologous to that of the AIF-1 in human GenBankrHSU49392 , pig Swiss
. Ž . Ž .
protrAIF1 PIG , mouse GenBankrAB013745 , rat GenBankrRNU17919 , and carp–
Ž .
Fig. 3. Nucleotide sequence and structure of protein coding region in red sea bream AIF-1 gene. A The six open reading frames are underlined, and the deduced amino acid sequence is aligned. The three asterisks
X X Ž .
Table 1
Similarities of the AIF-1 chains in red sea bream, carp, and mammals
RS Carp Mouse Rat Pig
Carp 59.9
Mouse 65.5 59.5
Rat 66.9 62.2 93.9
Pig 65.5 62.2 85.7 87.1
Human 65.5 63.5 89.1 89.8 91.2
Ž . Ž .
Red sea bream RS identical matches % .
ŽGenBankrAB012309 . The amino acid sequence from red sea bream AIF-1 were found. to be 65.5% homologous to those from human AIF-1, 65.5% homologous to those from pig AIF-1, 66.9% homologous to those from rat AIF-1, 65.5% homologous to those
Ž .
from mouse AIF-1 and 59.9% homologous to those from carp Table 1 .
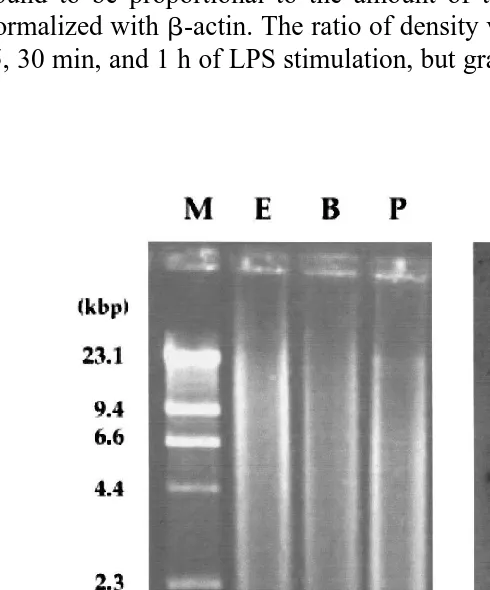
3.2. Southern blot analysis of red sea bream genomic DNA
Southern blot analysis of genomic DNA was performed using an AIF-1 gene
Ž .
fragment 1343 bp of PCR product; see Fig. 3 as a hybridization probe. As shown in
Fig. 4. Comparison of red sea bream AIF-1 chain with those of carp and mammals. Dashes indicate the amino acid gaps that are necessary to align these sequences. The conserved residues in all sequences are indicated by
Ž .
Fig. 4, a 4.4-kb long positive band was observed in the EcoRI lane Fig. 5 , and an additional faint band was observed just above the 4.4-kb band. In the lanes of BamHI and Pst I digestion, a 3.1- and 1.8-kb long single positive band was observed,
respec-Ž .
tively Fig. 5 . However, there were no faint bands in the lanes of BamHI and Pst I
Ž .
digestion Fig. 5 .
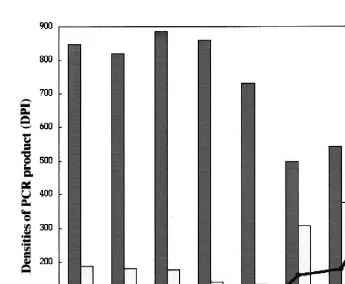
3.3. Time course of AIF-1 expression in LPS-stimulated leukocytes
From the calibration assay, both the density of PCR products of b-actin or AIF-1
Ž .
were found to be proportional to the amount of template cDNA Fig. 1 . The assays
Ž .
were normalized withb-actin. The ratio of density values AIF-1rb-actin were static at 0, 5, 15, 30 min, and 1 h of LPS stimulation, but gradually increased from 3 to 24 h. The
Fig. 5. Southern blot analysis of the red sea bream AIF-1 gene. Red sea bream genomic DNA was digested
Ž . Ž . Ž .
with EcoRI lanes E , BamHI lanes B , and Pst I lanes B . The digests were separated on 0.6% agarose gel
Žleft panel , blotted onto a nylon membrane and hybridized with radiolabeled probes right panel . Marker. Ž .
Ž . Ž .
DNA HindIII-digestedl-DNA was electrophoresed in parallel lane M . The filters were hybridized with a
Ž .
Fig. 6. Time course of AIF-1 expression level in LPS-stimulated red sea bream leukocytes. Red sea bream
Ž 6 . Ž
leukocytes 5=10 cells were placed into a 12-well plate, and subjected to LPS stimulation final
.
concentration: 0.1 mgrml . The RNAs were reverse transcribed with oligo dT, and amplified with primers for b-actin and AIF-1, respectively. The PCR conditions were calibrated to be consistent with template quantity.
Ž . Ž . Ž .
The bars represent density values DPI of PCR products by AIF-1 white andb-actin gray primers. The line indicates estimated expression level of AIF-1 based on the expression level ofb-actin.
Ž maximum ratio of expression values in AIF-1rb-actin was 24 h of LPS stimulation Fig.
. 6 .
4. Discussion
In this paper, we demonstrated the complete nucleotide sequences of red sea bream AIF-1 cDNA, and the coding region of red sea bream AIF-1 gene. All of the 12 cDNA
Ž
clones encode exactly the same sequence. Three plasmid clones that encode PCR .
one genomic DNA in red sea bream. A 32P-labelled AIF-1 gene fragment was hybridized with 4.4-, 3.1-, and 1.8-kb fragments in the lanes of EcoRI, BamHI, and
Ž .
HindIII digests, respectively Fig. 5 . As the single positive band in each of the three
lanes suggests, the AIF-1 gene might involve a single copy per haploid in the red sea bream genome. A 6.1-kb long faint band in the EcoRI lane is interesting and requires further analysis. However, because of its low affinity for the probe, it is probably just
Ž .
another gene Fig. 5 .
In the determined coding region of the red sea bream AIF-1 gene, six ORFs and five
Ž .
IVSs were observed. The general construction six exons and five introns and
exon–in-Ž .
tron junction sites were similar to those of human AIF-1 Utans et al., 1996rHSY14768
Ž .
and mouse AIF-1 direct submission: MMU82792 . This indicates the stability of the
AIF-1 gene in vertebrate evolution. The results of the semi-quantitative RT-PCR assay
used to assess AIF-1 expression suggests that the response to LPS stimulation is not as Ž
acute as that of cytokines. Since the rat AIF-1 was induced by IFN-g Utans et al., .
1995 , the lack of a response in red sea bream during the first 3 h may be due to a reaction time for activation of a cytokine network cascade. The parallel characteristics of AIF-1s in mammals and red sea bream suggest that the AIF-1 in red sea bream may have a similar function as AIF-1s in mammals.
Acknowledgements
Ž .
This study has been supported by Research for the Future Program RFTF by the
Ž .
Japan Society for the Promotion of Science JSPS . Project number: 97L00902.
References
Autieri, M.V., 1996. cDNA cloning of human allograft inflammatory factor-1: tissue distribution, cytokine induction, and mRNA expression in injured rat carotid arteries. Biochem. Biophys. Res. Commun. 228, 29–37.
Autieri, M.V., Feuerstein, G.Z., Yue, T.L., Ohlstein, E.H., Douglas, S.A., 1995. Use of differential display to identify differentially expressed mRNAs induced by rat carotid artery balloon angioplasty. Lab. Invest. 72, 656–661.
Chen, Z.W., Ahren, B., Ostenson, C.G., Cintra, A., Bergman, T., Moller, C., Fuxe, K., Mutt, V., Jornvall, H.,
Ž
Efendic, S., 1997. Identification, isolation, and characterization of daintain allograft inflammatory factor
.
1 , a macrophage polypeptide with effects on insulin secretion and abundantly present in the pancreas of pre-diabetic BB rats. Proc. Natl. Acad. Sci. U. S. A. 94, 13879–13884.
Chomczynski, P., Sacchi, N., 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate– phenol–chloroform extraction. Anal. Biochem. 162, 156–159.
Negrutskii, B.S., El’skaya, A.V., 1998. Eukaryotic translation elongation factor 1 alpha: structure, expression, functions, and possible role in aminoacyl-tRNA channeling. Prog. Nucleic Acid Res. Mol. Biol. 60, 47–78. Sambrook, J., Fritsch, E.F., Maniatis, T., 1989. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor
Laboratory Press, New York.
Utans, U., Arceci, R.J., Yamashita, Y., Russell, M.E., 1995. Cloning and characterization of allograft inflammatory factor-1: a novel macrophage factor identified in rat cardiac allografts with chronic rejection. J. Clin. Invest. 95, 2954–2962.
Utans, U., Quist, W.C., McManus, B.M., Wilson, J.E., Arceci, R.J., Wallace, A.F., Russell, M.E., 1996. Allograft inflammatory factory-1. A cytokine-responsive macrophage molecule expressed in transplanted human hearts. Transplantation 61, 1387–1392.