Cloning and in vitro expression of the cDNA encoding a putative
nucleoside transporter from
Arabidopsis thaliana
Jian Li, Daowen Wang *
The State Key Laboratory of Plant Cell and Chromosome Engineering,Institute of Genetics,The Chinese Academy of Sciences,
Beijing 100101, PR China
Received 8 February 2000; received in revised form 23 March 2000; accepted 24 March 2000
Abstract
Nucleoside transporters are integral membrane proteins involved in the uptake or release of nucleosides. Their function constitutes an essential step of the salvage pathway of nucleotide synthesis. In order to study the function of these proteins in higher plants, we cloned the cDNA corresponding to the AtENT1 gene that encodes a putative nucleoside transporter in
Arabidopsis thaliana by RT-PCR. The amino acid sequence of the AtENT1 protein deduced from the cloned cDNA shared similarity to those of eukaryotic equilibrative nucleoside transporters. Structure prediction indicated that the deduced AtENT1 protein might possess eleven putative transmembrane domains. Southern hybridization revealed thatAtENT1 had one homologue in the Arabidopsisgenome. Northern blot analysis showed that AtENT1 might be constitutively expressed in mostArabidopsis
organs and in plants at different developmental stages. TwoAtENT1 fusion genes, AtENT1-His-tag and GFP-AtENT1-His-tag, were expressed in insect cells. Confocal microscopy demonstrated that the GFP-AtENT1-His-tag fusion protein was targeted specifically to the plasma membrane of insect cells. © 2000 Elsevier Science Ireland Ltd. All rights reserved.
Keywords:Nucleoside transporter; cDNA cloning; In vitro expression;Arabidopsis thaliana
www.elsevier.com/locate/plantsci
1. Introduction
Considerable progress has been made in recent years in biochemical and molecular characteriza-tion of nucleoside transport (NT) processes in eukaryotic organisms such as mammals and proto-zoan parasites [1 – 9]. To date, seven types of NT processes have been described for mammalian cells [1,2]. The nucleoside transporters required for the different NT processes comprise two functionally different and structurally unrelated families of transmembrane proteins [1,2]. The equilibrative nucleoside transporters (ENTs) facilitate the diffu-sion of nucleosides down their concentration gra-dients [1 – 5]. According to their sensitivity to inhibition by nitrobenzylmercaptopurine ribonu-cleoside (NBMPR), the ENTs can be subdivided
into eiand estypes. Transport mediated by the ei
transporters is insensitive to NBMPR inhibition
whereas that mediated by the es transporters is
sensitive to inhibition by nanomolar concentra-tions of NBMPR. Nucleoside transport mediated by the ei and es transporters is bidirectional. The concentrative nucleoside transporters (CNTs) are Na+-dependent symporters [1 – 5]. They transport
nucleosides unidirectionally and against their con-centration gradients. Based on their selectivity, the CNTs can be further divided into five types: cit,
cif, cib, csgand cs. Nucleoside transport processes mediated by the five types of CNTs have recently been reviewed [1 – 5]. While the ENTs are present in most mammalian cell types, the CNTs are found primarily in specialized epithelia [1 – 5]. Some cell types can simultaneously possess several kinds of nucleoside transport activities, indicating the presence of multiple types of nucleoside trans-porters. Equilibrative nucleoside transport pro-* Corresponding author: Tel.: +86-10-64889380; fax: +
86-10-64873482.
E-mail address:[email protected] (D. Wang).
cesses and ENTs have been studied for protozoan
parasites such as Leishmania dono6ani, Try
-panosoma brucei and Toxoplasma gondii [6 – 9]. These parasites lack the de novo purine biosynthe-sis pathway and, consequently, have to rely on nucleoside transporters to acquire purine nu-cleosides from their host cells [10].
Genes encoding eukaryotic ENTs have been cloned from protozoan parasites and mammals [6 – 9,11 – 14]. Amino acid sequence comparisons suggest structural conservation of this type of genes between mammals and protozoan parasites. Genes encoding CNTs have mainly been isolated from mammalian cells [15 – 19]. A high degree of structural conservation is also found among the CNT encoding genes. Nucleoside transport func-tion has been confirmed for all of the cloned genes
through in vitro expression in Xenopus lae6is
oocytes followed by nucleoside uptake tests. More
recently, the Xenopus and the yeast expression
systems have been employed successfully for studying amino acid residues involved in nu-cleoside or inhibitor (NBMPR) binding in human CNTs (hCNTs) or ENTs (hENTs) [20,21]. The combination of molecular genetic and biochemical approaches is yielding new information on nu-cleoside transporters at a rapid pace. A deeper understanding of these proteins in mammalian and protozoan biology will become available before long.
In contrast to above progress, fewer investiga-tions have been carried out to characterize
nu-cleoside transporters in higher plants.
Physiological studies suggest that higher plants can salvage nucleosides and bases derived from nucleotide breakdown or from exogenous sources [22 – 25], indicating the existence and function of nucleoside transporters in plant cells. Plant nucleic acid sequences encoding potential polypeptides with homology to hENTs have recently been dis-covered in EST (expressed sequence tag) [26] and
genomic sequencing projects of Arabidopsis
thaliana. In the work described in this paper, we report our results on cloning and in vitro expres-sion of the cDNA encoding a putative nucleoside transporter from Arabidopsis thaliana. Our work presents molecular evidence for the expression of theAtENT1 gene in the Arabidopsis plant and for the association of the AtENT1 protein with the plasma membrane of eukaryotic cells.
2. Materials and methods
2.1. Plant growth and nucleic acid isolation
Arabidopsis thalianaecotype Columbia was used throughout this study. Seedlings germinated in composts were grown in the greenhouse at 20 –
22°C with supplemented light (14 h light/10 h
dark). Total RNA was isolated by using the Trizol reagent (Gibco BRL) according to the manufac-turer’s instruction. Genomic DNA was isolated using the CTAB method as described [27].
2.2. RT-PCR, cDNA cloning and DNA sequencing
Conceptual translation of four ESTs (EMBO
accessions N65317, T20785, AA586285,
AA712578) and one genomic (AAC18807,
PID:g3176684) Arabidopsis sequences yielded
polypeptides showing partial homology to hENT proteins. This suggests that, in the Arabidopsis
genome, the equilibrative nucleoside transporter
encoding gene (AtENT) has at least one member
that is transcribed. Because the genomic sequence contained regions showing complete nucleotide homology with the four EST sequences, we de-duced that the five sequences represent a single
gene, which we named as AtENT1. In order to
obtain the cDNA for the coding region of
AtENT1, two primers, NT1 (5%-CTAAACG
-GATCCAAATGACCACCAC-3%, the italicized nucleotides constituted a BamHI restriction site)
and NT2 (5%-CGAAGCTTAAAGAATTCAAC-3%,
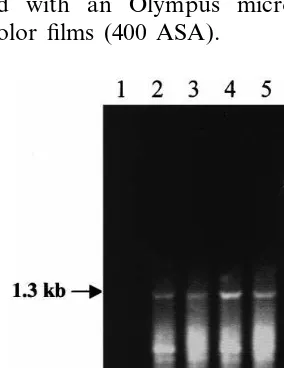
the italicized nucleotides constituted an EcoRI restriction site) were designed based on the avail-able sequence information. The primers were sub-sequently synthesized (by Gibco BRL) and used in RT-PCR to amplify a 1.3 kb DNA fragment
corresponding to the coding region of AtENT1.
pGEM-T vector (Promega). The insert in two independent clones was completely sequenced by a commercial company (TakaRa). One of the two clones was designated as pGEM-ANT and was used in the subsequent hybridization and cloning experiments.
2.3. Amino acid comparison and structure prediction
The cDNA sequence of AtENT1 coding region
was translated into amino acid sequence using the ORF Finder program (NCBI). The resultant amino acid sequence was compared with those of mammalian and protozoan ENTs using the Blast program (NCBI). The presence and arrangement
of transmembrane domains in AtENT1 protein
was predicted as described by Tusnady and Simon [28] and were subsequently compared with those of hENT1 and hENT2.
2.4. Southern and northern analysis
Genomic DNA from Arabidopsis (5 mg) was
digested with HindIII, EcoRI or DraI. The di-gested DNA was separated in 1% agarose gels.
After blotting onto the Hybond N+ membrane
(Amersham), DNA samples were hybridized with
32P-labelled probe prepared by priming AtENT1
cDNA (the insert in pGEM-ANT) using the
Prime-A-Gene® labeling system (Promega) [29].
Post hybridization washes used both low (2×
SSC/0.1% SDS, 65°C for 20 min each) and high
(0.1×SSC/0.1% SDS, 65°C for 20 min each) strin-gency procedures. Washed membranes were ex-posed to Kodak films and autoradiographies were obtained for all hybridized membranes.
To evaluate the expression of AtENT1 in Ara
-bidopsis thaliana, total RNA was extracted from different organs (root, leaf, rachis and flower) and from plants at eight developmental stages. The definition of the different developmental stages was as follows. Stage 1: cotyledons unfolded. Stage 2: second true leaf emerged. Stage 3: sixth true leaf emerged. Stage 4: rosette (no internodes). Stage 5: flower buds visible. Stage 6: flower buds raised above leaves. Stage 7: First flower buds white. Stage 8: 50% of all buds opened. Total
RNA samples (20 mg each) were run in
formalde-hyde-containing, denaturing agarose gels. Capil-lary transfer of RNA and blot hybridization with
32P-labeled probe (prepared as outlined above)
were carried out as described [29]. The hybridized blots were washed with both low and high strin-gency procedures. After an initial exposure for the
signal generated by AtENT1 mRNA, the blots
were stripped and were further hybridized with a probe specific for 18S rRNA to check the loading of different RNA samples during elec-trophoresis.
2.5. Expression in insect cells
Two fusion genes (AtENT1-His-tag and GFP-AtENT1-His-tag) were prepared for expression in insect cells using the Bac-to-Bac™ baculovirus expression system (Gibco BRL). To construct AtENT1-His-tag (the His-tag is a peptide sequence consisting of 6 histidines), two primers, NT1
(Sec-tion 2.2) and NT3 (5%-CAAATGAATTCTCAGT
-GATGGTGATGGTGATGAATGACCCAGAACC
AAGC-3%, the italicized nucleotides formed the
coding sequence for His-tag), were used to amplify
the coding region of AtENT1 from the
pGEM-ANT plasmid. The GFP (green fluorescence protein)-AtENT1-His-tag fusion was prepared in two steps. Firstly, a DNA fragment containing the coding region of GFP was amplified from
p35S-GFP plasmid [30] using primers G1 (5%
-
GATGGATCCATGAGTAAAGGAGAAGAAC-3%) and G2 (5%
-CTGGAGGATCCCTTTG-TATAGTTCATCC-3%). Secondly, the GFP
fragment was fused to the 5% end of the AtENT1-His-tag fragment using the restriction siteBamHI. In frame fusion between GFP and AtENT1-His-tag was confirmed by DNA sequencing. By follow-ing the instructions detailed by Gibco BRL, the above sequences were cloned downstream of the polyhedron promoter in the baculovirus genome. The resulted bacmids that contained the recombi-nant baculovirus genomes were introduced into insect cells (sf21). Expression of the two fusion genes in sf21 cells was checked either by immuno-cytochemical staining (for AtENT1-His-tag) or by confocal microscopy (for GFP-AtENT1-His-tag). In addition to the above bacmids specifying the
expression of AtENT1-His-tag and
2.6. Immunohistochemical staining and fluorescence microscopy
To visualize the expression of AtENT1-His-tag, sf21 cells at 48 h post transfection were collected by centrifugation (1000 g for 2 min), fixed with 4% paraformaldehyde (for 10 min) and spread onto poly-L-lysine (Sigma) treated slides. After 10 min
of drying on a warm plate (set at 45°C), the slides were washed with TBS (150 mM NaCl, 100 mM Tris – HCl, pH 7.5) for 5 min, followed by an
incubation in TBSM (TBS containing 5%
skimmed milk powder) for 60 min to block non-specific binding sites. After the incubation, the cells were treated with a mouse monoclonal anti-body that recognized the His-tag (Pharmacia, 1:2000 dilution in TBSM) for 24 h at 4°C. The antibody reaction was terminated by washing the cells three times (5 min each) in TBS. A goat anti-mouse IgG-alkaline phosphatase conjugate solution (Sigma, 1:2000 dilution in TBS) was then applied to the washed cells. After a 1-h reaction, the cells were washed three times with TBS (5 min for each wash). To detect antibody/conjugate reac-tion, the cells were treated with a substrate solu-tion containing NBT and BCIP as detailed by Wang and Maule [31]. To check the specificity of the monoclonal antibody, sf21 cells transfected with the WT bacmid were stained exactly as de-scribed above. The staining results were pho-tographed with an Olympus microscope using Kodak color films (400 ASA).
For visualizing the expression of GFP-AtENT1-His-tag by fluorescence microscopy, cells at 48 h post transfection were collected and spread onto clean slides. The cells were mounted in 10%
glyc-erol (in 130 mM NaCl, 7 mM Na2HPO4, 3 mM
NaH2PO4) containing 1 mg/ml propium iodide
(Sigma) and immediately examined under either a conventional fluorescence microscope (Olympus) or a confocal microscope (Bio-Rad 680) using appropriate excitation and emission filters. In con-focal microscopy, selected cells were optically sec-tioned at a 2 mm interval. On average, about 15
sections were obtained for each of the
examined cells. Fluorescent signals in all sections were carefully examined and recorded. Cells trans-fected with the P1-GFP bacmid were also exam-ined via confocal microscopy. The GFP fusion protein specified by this bacmid was used as a positive control for observing GFP fluorescence in insect cells.
3. Results
3.1. Cloning and amino acid sequence analysis of AtENT1 cDNA
RT-PCR reactions with primers NT1 and NT2 generated a cDNA fragment of approximately 1.3 kb (Fig. 1). The size of this fragment corresponded well with that of the AtENT1 coding region calcu-lated from the genomic sequence. Using the T/A cloning strategy, the PCR fragment was cloned into the pGEM-T vector. The cDNA sequence of the insert in two independent clones was exactly the same, and in both sequences a single ORF coding for a polypeptide of 428 amino acids was identified. Although equal in the number of the amino acids, the AtENT1 polypeptide specified by the cloned cDNA differed from the one deduced from the genomic sequence at two amino acid positions owing to two single nucleotide substitu-tions. In positions 89 and 410 of the polypeptide specified by the cDNA, a proline and a threonine residue were present, respectively, whereas in the polypeptide deduced from the genomic sequence the corresponding positions were occupied by a leucine and a methionine residue, respectively. The
amino acid sequence predicted from AtENT1
cDNA possessed significant similarity to those of several ENTs. Pairwise comparisons revealed that the putative nucleoside transporter AtENT1 was
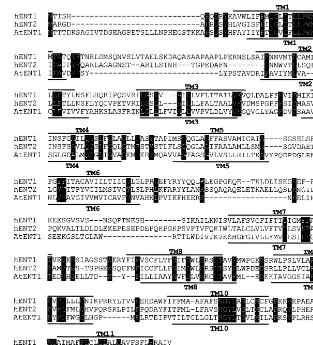
Fig. 2. Comparison of the deduced amino acid sequences of hENT1, hENT2 and the putative nucleoside transporter AtENT1. The AtENT1 amino acid sequence was derived by conceptual translation of theAtENT1 cDNA sequence. The three sequences were aligned using programs of the NCBI Network Services. Identical amino acids in the three sequences are shown in white on a black background. Solid lines above hENT1 and below AtENT1 sequences indicate putative transmembrane (TM) domains that may be present in the two proteins. The numbers on the left and right of the figure are amino acid positions in each sequence.
24% identical to hENT1, 25% to hENT2, 24% to rENT1 (rat equilibrative nucleoside transporter 1),
26% to rENT2, 22% to LdNT1 (Leishmania dono
-6ani nucleoside transporter 1) and 21% to TbNT2
(Trypanosoma brucei nucleoside transporter 2). In
addition to above ENTs, two hypothetical Ara
-bidopsis proteins (AAD25545 and AAF04424) identified by the genome-sequencing project also showed similarity to AtENT1 (28% identity was found between AAD25545 and AtENT1 and 29%
between AAF04424 and AtENT1). Eleven
transmembrane domains were predicted for the putative nucleoside transporter AtENT1. The or-ganization of the putative transmembrane do-mains in AtENT1 was similar to that in hENT1 (Fig. 2).
3.2. Detection of AtENT1 related DNA sequence in Arabidopsis genome
Genomic DNA was extracted from Arabidopsis
plants and was digested with several restriction enzymes. The gel blots were hybridized with 32
P-labelled probes. Post-hybridization washes used both high and low stringency washing conditions. Fig. 3 showed a typical example of the autoradio-graphies obtained. Two hybridizing bands were
present in EcoRI digested DNA (Fig. 3). Three
hybridizing bands were found for HindIII and
DraI digested DNA, but the two smallest bands
only one additional sequence (gene) that was re-lated to AtENT1 gene.
3.3. Expression of AtENT1 mRNA in different organs and in plants at different de6elopmental stages
Two sets of Northern blot hybridization experi-ments were performed to investigate the expres-sion of AtENT1 in Arabidopsis. The intensity of the hybridization signals generated from total RNA samples extracted from leaf, root, rachis and flower of fully-grown Arabidopsis plants was of a similar level (Fig. 4A). When total RNA samples derived from plants at eight different developmen-tal stages were hybridized, there was also little difference in the hybridization signal in between the eight samples (Fig. 4B).
3.4. In 6itro expression of AtENT1 fusion genes in insect cells
For future functional study, it is important to
test the expression of AtENT1 in an eukaryotic
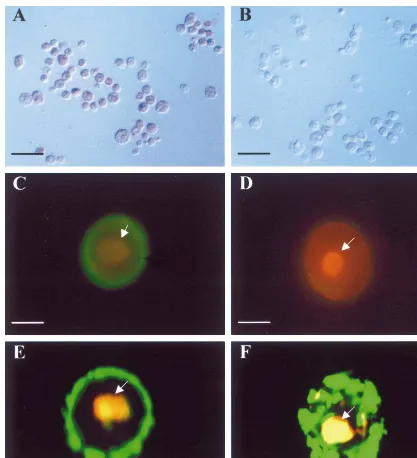
expression system and to obtain information on intracellular localization of the putative nucleoside transporter AtENT1 in eukaryotic cells. For this purpose, we expressed the AtENT1-His-tag fusion gene in insect cells. By immunocytochemical stain-ing usstain-ing a monoclonal antibody specific for the His-tag, positive expression of the AtENT1-His-tag fusion gene in sf21 cells was demonstrated (Fig. 5A and B). Although the signal produced by immunocytochemical staining was sufficient for checking fusion gene expression, it was not condu-cive for examining intracellular localization of the expressed product. To overcome this problem, a second fusion gene, prepared by adding the GFP
coding region to the 5% end of AtENT1-His-tag,
was expressed in sf21 cells. When observed under a conventional fluorescence microscope, 30 – 50% of the cells exhibited GFP fluorescence and the GFP fluorescence was associated with the plasma membrane (Fig. 5C). No GFP fluorescence was detected in the cells expressing the WT type genome of the baculovirus (Fig. 5D). Expressing cells were further examined under a confocal mi-croscope, because we wished to investigate if there was also some GFP fluorescence present in intra-cellular membrane systems (such as endoplasmic reticulum and nuclear membrane). Optical section-ing permitted a more detailed, and hence more precise, localization of GFP fluorescence within the entire contents of the examined cells. The results showed that, in the cells expressing GFP-AtENT1-His-tag, GFP fluorescence was localized
Fig. 3. Detection ofAtENT1 related DNA sequence inAra
-bidopsisgenome by Southern blot hybridization. Aliquots (5
mg each) of genomic DNA were digested withEcoRI (lane E),
HindIII (lane H) orDraI (lane D), the digested DNA samples were fractionated on 1% agarose gels followed by transfer onto Hybond N+ membrane. The resulted blots were
hy-bridized with 32P-labelled probes prepared with AtENT1
cDNA. The size of DNA markers is indicated in kilobase (kb) pairs. The two bands indicated by arrow are too small to represent intact genes.
Fig. 4. Northern hybridization analysis ofAtENT1 mRNA in different organs of fully-grownArabidopsisplants (A) and in
Arabidopsisplants at different developmental stages (B). Total RNA samples (20mg each) from root (lane RO), rachis (lane
RA), leaf (lane LE), flower (lane FL), plants at four vegeta-tive stages (lanes 1 – 4) and plants at four reproducvegeta-tive stages (lane 5 – 8) were fractionated on agarose gels prior to transfer to Nylon membrane. The resultant blots were hybridized firstly with32P-labelled probes prepared withAtENT1 cDNA
to detect AtENT1 mRNA. After stripping, the blots were further hybridized with 32P-labelled probes specific for 18S
Fig. 5. Expression of twoAtENT1 fusion genes in insect cells. AtENT1-His-tag expressing sf 21 cells (A) and the cells expressing wild type baculovirus genome (B) were immuno-stained with an anti-His-tag monoclonal antibody. The cells shown in A were positively stained. In (C), a sf21 cell expressing GFP-AtENT1-His-tag was examined and photographed with a conventional fluorescent microscope, GFP fluorescence (green color) was associated with the cell periphery. No GFP fluorescence was observed in the sf21 cell expressing wild type baculovirus genome (D). In (E), a confocal image of a sf21 cell expressing GFP-AtENT1-His-tag is shown. GFP fluorescence was associated specifically with the plasma membrane. In contrast, in the cell expressing a predominantly cytosolic GFP fusion protein (F), GFP fluorescence was distributed mainly in the cytoplasm (F). In (C – F), cells were counter-stained with a PI solution to reveal the nuclei (arrowed). Bars represent 60mm (in A and B), 15mm (in C and D)
and 10mm (in E and F).
to the plasma membrane (Fig. 5E) and no GFP fluorescence was detected to be associated with other cellular constituents. In contrast, in the cells expressing the cytosolic P1-GFP fusion protein, GFP fluorescence resided predominately in the cytoplasm (Fig. 5F).
4. Discussions
expression of the AtENT1 gene in the Arabidopsis
plant was also investigated using Northern hy-bridization assays. The availability of four EST and one genomic sequences for AtENT1 facilitated the design of nucleotide primers for RT-PCR reactions. The cloning and subsequent sequencing of the PCR generated cDNA fragment confirmed the presence inArabidopsisof a mRNA capable of directing the synthesis of the AtENT1 polypeptide originally deduced from the genomic sequence.
Amino acid sequence analysis indicated that ENTs from mammals and protozoan parasites were structurally related, with 20 – 30% of identity in between the different polypeptides [6 – 9]. Our work showed that the deduced polypeptide of the putative nucleoside transporter AtENT1 was 20 – 30% iden-tical to those of mammalian and protozoan ENTs and that the AtENT1 and hENT1 polypeptides may possess an identical number of putative transmem-brane domains. These results provided further evi-dence that ENTs encoded by the nuclear genome of different eukaryotic organisms may constitute a family of structurally and functionally related inte-gral membrane proteins.
Based on the pattern of hybridizing bands in Southern blot experiments, we deduced that
AtENT1 had only one more homologous gene in the
Arabidopsis genome. The sequences encoding the two hypothetical proteins that showed 28 – 29%
identity to AtENT1 in amino acid sequence were
unlikely to produce hybridization signals in our Southern analysis because their homology to
AtENT1 at the nucleotide level was too low. The interpretation of the results from our Northern blot analysis may be complicated by the presence of an
AtENT1 homologue in theArabidopsisgenome. If theAtENT1 homologue was expressed, the North-ern hybridization signal shown in Fig. 4 would be produced by transcripts from two related genes. If the AtENT1 homologue did not express or ex-pressed only at a negligible level under normal
growth condition, we may conclude that AtENT1
possessed a pattern of constitutive expression. Be-fore we determine the expression pattern of the
AtENT1 homologue, the conclusion on AtENT1 expression in this study could only be tentative. But, no matter what is the final conclusion, it is clear that the AtENT1 polypeptide, and possibly its ho-mologous protein as well, may function throughout the entire life cycle of the Arabidopsis plants.
Our attempts in expressingAtENT1 gene in insect cells provided two pieces of important information. First, the baculovirus-insect cell expression system allows the translation of this type of plant trans-porters. Second, the translated products of the fusion genes such as GFP-AtENT1-His-tag are correctly targeted to the plasma membrane, which proves the function of the putative nucleoside transporter AtENT1 as a plasmalemma located
transporter. The application of GFP and AtENT1
fusion constructs should greatly facilitate future investigations in determining cell type specific ex-pression ofAtENT1 and the function of the protein in Arabidopsis biology. However, the nucleoside transport function of the proteins specified by any GFP-AtENT1 fusion genes must be determined first. We originally thought to useAtENT1 express-ing insect cells to conduct nucleoside uptake exper-iments. But we were discouraged by the finding that insect cells possessed multiple types of endogenous NT activities [32]. Recently, the yeast expression system has been successfully employed to study nucleoside transport function of hENTs [21]. The adoption of yeast cells, as an alternative het-erologous expression system for testing nucleoside transport function will be vigorously pursued in the near future.
Acknowledgements
The authors wish to thank Dr Georg Leggewie (MPI fuer Molekulare Pflanzenphysiologie, Golm, Germany) for constructive discussions and for cor-recting the English. Dr Andy Maule (John Innes Centre, UK) is thanked for providing the bacmid specifying the expression of the P1-GFP fusion protein in insect cells. In addition, we are grateful to Dr Y. Xu (Texas A&M University) for providing the plasmid that was used for preparing 18S rRNA specific probes in this investigation. This work was supported by the Chinese National Nature Science Foundation (grant 39770491).
References
[2] C.E. Cass, J.D. Young, S.A. Baldwin, Recent advances in the molecular biology of nucleoside transporters of mammalian cells, Biochem. Cell Biol. 76 (1998) 761 – 770.
[3] J.S. Wiley, Seeking the nucleoside transporter, Nat. Med. 3 (1997) 25 – 26.
[4] J.A. Thorn, S.M. Jarvis, Adenosine transporters, Gen. Pharmacol. 27 (1996) 613 – 620.
[5] D.A. Griffith, S.M. Jarvis, Nucleoside and nucleobase transport systems of mammalian cells, Biochem. Bio-phys. Acta 1286 (1996) 153 – 181.
[6] G. Vasudevan, N.S. Carter, M.E. Drew, S.M. Beverley, M.A. Sanchez, A. Seyfang, B. Ullman, S.M. Landfear, Cloning of Leishmania nucleoside transporter genes by rescue of a transport-deficient mutant, Proc. Natl. Acad. Sci. USA 95 (1998) 9873 – 9878.
[7] P. Ma¨ser, C. Su¨tterlin, A. Kralli, R. Kaminsky, A nu-cleoside transporter from Trypanosoma brucei involved in drug resistance, Science 285 (1999) 242 – 244.
[8] M.A. Sanchez, B. Ullman, S.M. Landfear, N.S. Carter, Cloning and functional expression of a gene encoding a P1 type nucleoside transporter fromTrypanosoma brucei, J. Biol. Chem. 274 (1999) 30244 – 30249.
[9] C.-W. Chiang, N. Carter, W.J. Sullivan Jr., R.G.K. Donald, D.S. Roos, F.N.M. Naguib, M.H. el Kouni, B. Ullman, C.M. Wilson, The Adenosine transporter of
Toxoplasma gondii, identification by insertional mutagen-esis, cloning, and recombinant expression, J. Biol. Chem. 274 (1999) 35255 – 35261.
[10] C.R. Crawford, D.H. Patel, C. Naeve, J.A. Belt, Cloning of the human equilibritive, nitrobenzylmercaptopurine riboside (NBMPR)-insensitive nucleoside transporter ei by functional expression in a transport-deficient cell line, J. Biol. Chem. 273 (1998) 5288 – 5293.
[11] M. Griffiths, S.Y.M. Yao, F. Abidi, S.E.V. Phillips, C.E. Cass, J.D. Young, S.A. Baldwin, Molecular cloning and characterization of a nitrobenzylthioinosine-insensitive (ei) equilibrative nucleoside transporter from human pla-centa, Biochem. J. 328 (1997) 739 – 743.
[12] M. Griffiths, N. Beaumont, S.Y. Yao, M. Sundaram, C.E. Boumah, A. Davis, F.Y. Kwong, I. Coe, C.E. Cass, J.D. Young, S.A. Baldwin, Cloning of a nucleoside transporter implicated in the cellular uptake of adenosine and chemotherapeutic drugs, Nat. Med. 3 (1997) 89 – 93. [13] S.Y. Yao, A.M. Ng, W.R. Muzyka, M. Griffiths, C.E. Cass, S.A. Baldwin, J.D. Young, Molecular cloning and functional characterization of nitrobenzylthioinosine (NBMPR)-sensitive (es) and NBMPR-insensitive (ei) equilibrative nucleoside transporter proteins (rENT1 and rENT2) from rat tissues, J. Biol. Chem. 272 (1997) 28423 – 28430.
[14] A. Felipe, R. Valdes, B. delSanto, J. Lloberas, J. Casado, M. PastorAnglata, Na+-dependent nucleoside
trans-porter in liver: two different isoforms from the same gene family are expressed in liver cells, Biochem. J. 330 (1998) 997 – 1001.
[15] M.W. Ritzel, S.Y.M. Yao, M.Y. Huang, J.F. Elliott, C.E. Cass, J.D. Young, Molecular cloning and func-tional expression of cDNAs encoding a human Na+
-de-pendent nucleoside cotransporter (hCNT1), Am. J. Physiol. 272 (1997) C707 – C714.
[16] J. Wang, S.-F. Su, M.J. Dresser, M.E. Schnaner, C.B. Washington, K.M. Giacomini, Na+-dependent purine
nucleoside transporter from human kidney: cloning and functional characterization, Am. J. Physiol. 273 (1997) F1058 – F1065.
[17] S.Y. Yao, A.M. Ng, M.W. Ritzel, W.P. Gati, C.E. Cass, J.D. Young, Transport of adenosine by recombinant purine- and pyrimidine-selective sodium/nucleoside co-transporters from rat jejunum expressed in Xenopus lae6isoocytes, Mol. Pharmacol. 50 (1996) 1529 – 1535.
[18] Q.Q. Huang, S.Y. Yao, M.W. Ritzel, A.R. Paterson, C.E. Cass, J.D. Young, Cloning and functional expres-sion of a complementary DNA encoding a mammalian nucleoside transport protein, J. Biol. Chem. 269 (1994) 17757 – 17760.
[19] M.E. Schaner, J. Wang, L. Zhang, S.F. Su, K.M. Ger-stin, K.M. Giacomini, Functional characterization of a human purine-selective, Na+-dependent nucleoside
transporter (hSPNT1) in a mammalian expression sys-tem, J. Pharmacol. Exp. Ther. 289 (1999) 1487 – 1491. [20] S.K. Loewen, A.M.L. Ng, S.Y.M. Yao, C.E. Cass, S.A.
Baldwin, J.D. Young, Identification of amino acid residues responsible for the pyrimidine and purine nu-cleoside specificities of human concentrative Na+
-depen-dent nucleoside cotransporters hCNT1 and hCNT2, J. Biol. Chem. 274 (1999) 24475 – 24484.
[21] M. F. Vickers, R.S. Mani, M. Sundaram, D.L. Hogue, J.D. Young, S.A. Baldwin, C.E. Cass, Functional pro-duction and reconstitution of the human equilibrative nucleoside transporter (hENT1) in Saccharomyces cere
-6isiae, interaction of inhibitors of nucleoside transport
with recombinant hENT1 and a glycosylation-defective derivative (hENT1/N48Q), Biochem. J. 339 (1999) 21 – 32.
[22] C. Wasternack, Uptake and incorporation of pyrimidines inEuglena gracilis, Arch. Microbiol. 109 (1976) 167 – 174. [23] A. Guranowski, J. Pawelkiewicz, Adenosylhomocys-teinase and adenosine nucleosidase activities in Lupinus luteus cotyledons during seed formation and germina-tion, Planta 139 (1978) 245 – 247.
[24] A. Guranowski, J. Barankiewicz, Purine salvage in cotyledons of germinating lupin seeds, FEBS Lett. 104 (1979) 95 – 98.
[25] A. Guranowski, C. Wasternack, Adenine and adenosine metabolizing enzymes in cell-free extracts of Euglena gracilis, Comp. Biochem. Physiol. 71B (1982) 483 – 488. [26] T. Newman, F.J. de Bruijn, P. Green, K. Keegstra, H.
Kende, L. McIntosh, J. Ohlrogge, N. Raikhel, S. Somerville, M. Thomashow, E. Retzel, C. Somerville, Genes galore: a summary of methods for assessing results from large-scale partial sequencing of anonymous Ara
-bidopsiscDNA clones, Plant Physiol. 106 (1994) 1241 – 1255.
[27] S.O. Rogers, A.J. Bendich, in: S.B. Gelvin, R.A. Schilperoort (Eds.), Plant DNA extraction, Plant Molec-ular Biology, vol. A6, Kluwer Academic, Dordrecht, Belgium, 1988, pp. 1 – 10.
[29] J. Sambrook, E.F. Fritsch, T. Maniatis, Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Laboratory, Cold Spring Harbor, New York, 1989. [30] J. Haseloff, K.R. Siemering, D.C. Prasher, S. Hodge,
Removal of a cryptic intron and subcellular localization of green fluorescence protein are required to mark trans-genicArabidopsis plants brightly, Proc. Natl. Acad. Sci. USA 94 (1997) 2122 – 2127.
[31] D. Wang, A.J. Maule, A model for seed transmission of a plant virus: genetic and structural analysis of pea embryo invasion by pea seed-borne mosaic virus, Plant Cell 6 (1994) 777 – 787.
[32] D.L. Hogue, C.E. Cass,Spodoptera frugiperda(Sf9) cells express novel purine- and pyrimidine-selective nucleoside transporters, Insect Biochem. Mol. Biol. 24 (1994) 517 – 523.