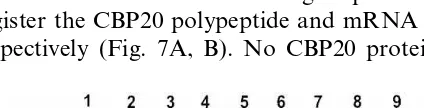Expression of the tobacco gene CBP20 in response to
developmental stage, wounding, salicylic acid and heavy metals
Go¨tz Hensel, Gotthard Kunze *, Irene Kunze
1Institut fu¨r Pflanzengenetik und Kulturpflanzenforschung(IPK),Corrensstr.3,D-06466Gatersleben,Germany
Received 23 April 1999; received in revised form 6 July 1999; accepted 12 July 1999
Abstract
The pathogenesis-related (PR)-protein CBP20 which is a basic representative of the group PR-4 is similarly regulated as the basic protein class-I b-1,3-glucanase and class I chitinase of PR-2 and PR-3, respectively. It shows complex hormonal, developmental and pathogenesis-related regulation. The protein is constitutively accumulated in suspension-cultured cells, in roots and in lower source leaves of healthy tobacco plants, but not in upper, sink leaves near the top of the plant. After spraying the plants with salicylic acid, the protein CBP20 was also detected in sink leaves, indicating a potential function of salicylic acid in the signal transduction pathway. Synthesis of CBP20 is inducible by heavy metals, especially zinc chloride, which was shown by Northern and Western blotting. We have demonstrated that the accumulation of mRNA not always resulted in accumulation of the appropriate protein leading to the suggestion that expression of CBP20 is transcriptionally as well as post-transcriptionally regulated by more than one response pathway. © 1999 Published by Elsevier Science Ireland Ltd. All rights reserved.
Keywords:Heavy metal ions;Nicotiana tabacum; Pathogenesis-related protein; Salicylic acid
www.elsevier.com/locate/plantsci
1. Introduction
Pathogenesis-related (PR)-proteins of plants are accumulated in response to infections by viruses, bacteria or fungi (for reviews, see [1 – 8]). In addi-tion, there are some members of different PR-families which have been found to be synthesized after treatments with chemicals such as air pollu-tants, phytohormones like auxins, cytokinins and ethylene, osmotic stress or salt stress conditions and others (for review, see [9]). Production of PR-proteins has also been shown to be develop-mentally regulated in healthy plants since high levels of some PR-proteins have been reported in roots, senescent leaves and during flowering [9]. The PR-protein CBP20 is known as a pathogen-and wound-inducible antifungal protein of 20
kDa firstly purified from tobacco (Nicotiana tabacum) Samsun NN leaves inoculated with to-bacco mosaic virus (TMV) [10]. It belongs to class I PR-4 proteins based on its structural and im-munological relationship to class II PR-proteins PR-4a and PR-4b of tobacco and PR-2 of tomato and its intracellular localization [10]. The authors demonstrated its accumulation by TMV, ethephon-treatment, wounding and UV light-treatment by Northern blotting analysis. But so far it is un-known, whether the increased transcript levels cause in each case an increased protein level as well. In this paper, we demonstrate that several envi-ronmental signals and different developmental stages induce the accumulation of CBP20 mRNA and protein. However, the mRNA level does not correlate in each case with the appropriate protein level and vice versa. In addition, a correlation between the levels of transcript and protein caused by heavy metals indicates at least two separate signal transduction pathways for the induction of CBP20.
* Corresponding author. Tel.: +49-394-825-520; fax: + 49-394-825-366.
E-mail address:[email protected] (G. Kunze)
1Present address: SunGene GmbH & Co. KG aA, Corrensstr. 3,
D-06466 Gatersleben, Germany.
2. Materials and methods
2.1. Plant material
N. tabacum L. cv. Samsun NN plants were grown in soil in a greenhouse at 18 – 22°C and 40 – 60% relative humidity. In vitro plants were cultured in Murashige and Skoog [11] medium without growth regulators and with 2% sucrose under a light – dark regime of 16:8 h at 3000 lux and a mean temperature of 24°C.
The cell suspension line 425 was started from callus of S275N, a suspension-cultured line of cloned N. tabacum L. cv. Havana 425 pith tissue [12,13], which was kindly provided by F. Meins (Basel, Switzerland). The culture was initiated and maintained as described previously [14].
2.2. Induction of CBP20
Greenhouse plants used for induction of CBP20 were 12 – 14 weeks old (3 weeks before
flow-ering). At this stage, they had 16 – 18 leaves. In vitro plants taken for the same kind of experi-ments were, if not otherwise noted, 4 – 6 weeks old with 8 – 10 leaves. Leaves were derived from ma-ture source to sink leaves as described in the figure legends. For wounding leaves, shoots and roots were scratched with razor blades arranged with a distance of 10 mm and subsequently placed in dishes in distilled water. The dishes were gently shaken on a gyratory shaker (60 rpm) in the dark at room temperature for different times as indicated.
For salicylic acid treatment, in vitro plants were sprayed with a solution of 50 mM and, after 48 h fully expanded leaves were harvested and frozen in liquid nitrogen.
Suspension-cultured cells were filtered and washed as described by Kunze et al. [14] and subsequently transferred into media containing different heavy metals in different concentrations as indicated in results. After incubation for differ-ent times, cells were harvested by filtration and extracted as described earlier [15].Each experiment was repeated at least twice.
2.3. Transcription inhibition experiments
Floating experiments with leaf disks were per-formed as described by Jacobs et al. [16] using
actinomycin D (Sigma, Germany) at 100 mg ml−1.
After incubation with and without actinomycin D for 2 and 4 h, leaves were used for RNA extrac-tion as described below.
2.4. RNA extraction and Northern blot experiments
Total RNA was prepared as described by Loge-mann et al. [17], separation of RNA in denaturing agarose gels, blotting and hybridization of nylon membranes were performed as described by Her-bers et al. [18]. The CBP20-containing fragment isolated by the Qiagen kit (Qiagen, Germany) from plasmid DNA of E. coli was used as probe.
2.5. Protein extraction and immunoblotting
Cells were collected on 56-mm pore-size nylon filters from suspension cultures after different in-cubation periods as indicated in figure legends. Extracts of cells, leaves, shoot segments and roots were prepared by freezing the material in liquid nitrogen, grinding with a mortar and pestle, and vortexing the ground material with five glass beads for 3 min in 1 ml buffer TNT (500 mM NaCl, 0.02% Triton X-100, 50 mM Tris-HCl, pH 8.0) per 1 g fresh weight of cells. After centrifugation (34,000 x g) for 15 min the supernatant was used to measure the protein concentration by the method of Bradford [19] using bovine serum albu-min (BSA) as a standard. Five mg protein per lane were separated by SDS-PAGE as described by Laemmli [20] except for using gradient gels from 11 to 20% polyacrylamide. Immunoblot analysis was performed as described elsewhere [21]. The antibody directed against CBP20 was prepared as described by Hensel et al. [15]. The antibody raised against the large subunit of the D
-ribulose-1,5-bisphosphate carboxylase (rubisco) was kindly provided by K.-H. Su¨ß (IPK, Gatersleben, Germany).
2.6. Chlorophyll assay
3. Results and discussion
The PR-4 family of proteins is organized simi-larly to that of the plant chitinase family (PR-3), in that both contain structural subclasses charac-terized by the presence or absence of an amino-ter-minal lectin domain. PR-4 genes have been described in Arabidopsis thaliana, Solanum tubero
-sum, N. tabacum, Lycopersicum esculentum and
He6ea brasiliensis [23 – 27], but a mature protein
has been characterized only from the monocot barley [28 – 30] and tobacco [10], which is the protein CBP20. The latter is 20 kDa in size and contains a chitin-binding domain with extensive similarity to the chitin-binding domains of chiti-nases [5], hevein [27], stinging nettle lectin [31] and the WIN proteins from potato [24]. Ponstein et al. [10] have tested the inducibility of CBP20 expres-sion on mRNA levels by different stress conditions like inoculation of leaf samples with TMV, ethep-hon treatment, wounding and UV light treatment. They concluded that the stress induction pattern of CBP20 matches the stress induction pattern of other class I PR-proteins. In this study we analysed the expression of CBP20 in response to senescence, wounding, salicylic acid and heavy metals on mRNA as well as protein level to obtain information about the expression pattern and its regulation.
3.1. The CBP20 gene expression is de6elopmentally regulated and induced by
senescence
Greenhouse plants (12 – 14 weeks old) of N.
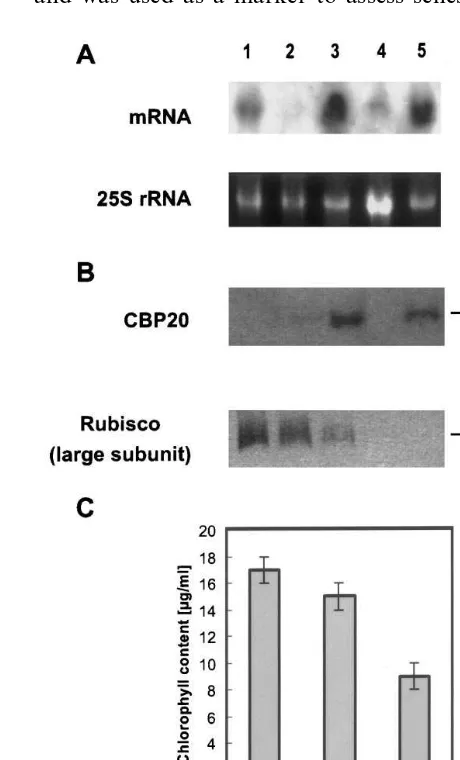
tabacum SNN were dissected into leaves, shoots and roots. Leaves were classified into old (1= old-est leaf), middle-aged and young leaves. Leaves 2+3, without visible signs of senescence, were used as old leaves. Leaves 8 and 9 were taken as middle-aged leaves and leaves 16 and 17 were used as young leaves. Leaves of the different stages, the shoot and roots were used for protein and RNA extraction as described in methods. Five mg protein of each fraction were loaded per lane, separated by SDS-PAGE and used for parallel immunoblotting to identify CBP20 (Fig. 1B). One polypeptide of 19 kDa could be detected in extracts of old leaves (lane 3) and roots (lane 5) of the greenhouse plant. This polypeptide corre-sponds to CBP20, because other immunologically
related proteins differ in molecular mass to CBP20. The accumulation of CBP20 in roots and lower leaves confirms the finding of Memelink et al. [32] that basic PR proteins are constitutively found in roots of healthy plants. In extracts of middle-aged leaves (lane 2) only a weak signal and in young leaves (lane 1) and shoots (lane 4) no signal was obtained with our anti-CBP20 antibody.
The protein rubisco is located in chloroplasts and was used as a marker to assess senescence of
leaves. The intensity of the polypeptide band cor-responding to the large subunit of rubisco de-creased with increasing age of leaves (lanes 1 – 3), which is a hint that senescence has already started (Fig. 1B). This was confirmed by the correlative chlorophyll levels (Fig. 1C). The increasing level of the CBP20 protein with increasing age of leaves on one hand and the starting senescence of these leaves on the other hand led us suggest that the expression of CBP20 was induced by senescence. To look if there is a correlation between the CBP20 expression on protein level as well as mRNA level Northern blotting experiments were performed. Twenty mg of total RNA prepared from the same explants used for Western blotting experiments, were loaded per lane and hybridized with the radiolabelled CBP20 cDNA (Fig. 1A). In case of old leaves (lane 3) and roots (lane 5) transcript and protein concentrations seemed to correlate with each other. However, the mRNA transcript level, of young leaves, middle-aged leaves and shoots (lanes 1, 2, 4) did not correlate with intensities of the appropriate CBP20 polypep-tide band. The mRNA signal of young leaves (lane 1) is more intensive than that of middle-aged leaves (lane 2), but on the protein level a weak signal was obtained for middle-aged leaves and no CBP20 protein was found in extracts of young leaves. We suggested therefore, that CBP20 is also post-transcriptionally regulated. This could be re-alized by different mRNA stabilities. It might be that the turnover of the CBP20 mRNA is higher in young leaves than in older ones with beginning senescence.
Leaf senescence is the final stage of leaf develop-ment and it is both highly regulated and geneti-cally programmed. Leaf cells experience sequential disorganization of cellular organelles and dramatic changes of cellular metabolism [33 – 36]. Leaf senescence is associated with induction of various genes which was shown in L. esculentum [37,38],
Brassica napus [39,40], A. thaliana [41,42], barley leaves [43], radish cotyledons [44] and others. Genes induced by senescence encode formerly degradative enzymes like proteases and nucleases [36,37].
Genes for senescence-specific proteases have been identified inA.thaliana [41,45] andZea mays
[35]. In addition to degradative enzymes the syn-thesis of PR-proteins is induced like that of chiti-nases of different classes [22] and PR-1 proteins
Fig. 2. Influence of wounding on the CBP20 mRNA level. Nonwounded (odd lane numbers) and wounded leaves (even lane numbers) were incubated for 0 (1,2); 1 (3,4); 4 (5,6) and 20 h (7,8) and subsequently used for Northern blotting.
[46,47]. InB.napusthe expression of a PR-1a gene and a gene for a class IV acidic chitinase is in-duced at different times during leaf development and becomes active in senescing leaves that are otherwise healthy [22].
3.2. Wounding induces an increased mRNA le6el
but not protein le6el
A missing correlation between the CBP20 mRNA level and the appropriate protein level was registered in case of wounding as well. Fully ex-panded leaves of a 9 weeks old in vitro plant were cut off. Leaves were wounded or not wounded, floated on distilled water for 0, 1, 4 and 20 h and subsequently used for Northern blotting (Fig. 2). A weak signal was registered in wounded as well as in nonwounded leaves immediately after wounding (lanes 1, 2). The intensity decreased after 1 h (lanes 3, 4) of incubation and increased again after 4 and 20 h (lanes 5 – 8). At these times a definite higher intensity was registered in wounded explants (even lane numbers) than in nonwounded leaves (odd lane numbers), which confirmed the results of Ponstein et al. [10] that the CBP20 expression is inducible by wounding at mRNA level. This also confirms the hypothesis of Brederode et al. [48], Bol et al. [8] and Pieterse and Van Loon [49] that wounding activates basic PR-proteins.
Fig. 3. Influence of wounding on the CBP20 protein level. Young leaves (1,4,7,10), shoots (2,5,8,11) and roots (3,6,9,12) were wounded and either immediately used for extraction (1 – 3) or subsequently floated on water for 1 (4 – 6), 4 (7 – 9) and 20 h (10 – 12).
SA-treatment CBP20 was additionally detected in young and middle-aged leaves (lanes 6, 7) which indicates an SA-inducible increase of the CBP20 protein level. In contrast, no increased protein levels were detected for basic chitinases after SA treatment (Fig. 4B, lanes 6, 7).
At least two separate pathways for pathogene-sis-inducible responses in tobacco are known. In the first SA which is known to induce systemic acquired resistance (SAR) strongly activates acidic PR genes as PR-1, PR-2 (acidic glucanases), PR-3 (acidic chitinases), PR-4 and PR-5 [55] whereas in the second pathway the basic representatives re-sponse to ethylene [56] and only weakly to SA. Expression analysis of a hevein-like (HEL) protein belonging to PR-4 like proteins revealed that accu-mulation of this HEL mRNA is strongly induced by ethylene treatment and is induced to a lesser extent by SA [57]. Classification into these two pathogenesis-responsible pathways are based on quantitative differences in accumulation of mRNA, whereas the appropriate protein levels were almost not considered. The accumulation of CBP20 as basic representative of the PR-4 class is also strongly inducible by ethephon [10] like basic chitinases. However, it differs by its additional inducibility by SA. Therefore, it should be as-sumed that CBP20 uses most likely another kind of signalling.
3.4. CBP20 is accumulated after incubation with hea6y metals
The influence of heavy metals was tested on the expression of CBP20 in suspension cells and leaves of in vitro plants. Suspension cells were trans-ferred 6 days after subculturing into medium sup-plemented with 100 mM of AgNO3, CdSO4, CoCl2,
CuSO4 and ZnCl2 and incubated for 15 h.
Subsequently, they were harvested, extracted as described in methods and used for Western blot-ting experiments (Fig. 5). In each case, one polypeptide of 19 kDa was detected with the anti-CBP20 antibody. The intensities of all CBP20 polypeptide bands were similar, except for the band of the ZnCl2-treated sample which was a
little bit more intensive (lane 6). To test the poten-tial influence of ZnCl2 on the inducibility of
CBP20 we observed the mRNA levels of suspen-sion cells during their incubation. Cells were trans-ferred 10 days after subculturing into medium with (lanes 1, 4, 7, 10) and shoots (lanes 2, 5, 8, 11) no
CBP20 protein was found after wounding up to 20 h of incubation. This indicates that wounding did not lead to an increased CBP20 protein level. This result confirms that the expression of CBP20 seems to be transcriptionally as well as post-tran-scriptionally regulated.
3.3. The accumulation of CBP20 is induced by salicylic acid (SA)
SA plays an important role in signal transduc-tion pathways [50 – 53]. Exogenously applied SA also results in an accumulation of some PR-proteins [8,52,54]. We tested the influence of ex-ogenously applicated SA on the accumulation of CBP20 and basic chitinase isoforms in in vitro plants. After SA-treatment as described in Section 2, 9 week old plants were dissected in old (leaves 1 and 2), middle-aged (leaves 4 and 5) and young (leaves 9 and 10) leaves, shoots and roots, ex-tracted and used for Western blotting experiments to register the CBP20 and chitinase protein levels (Fig. 4A, B). As expected, in nontreated plants CBP20 was found in extracts of roots and old leaves (Fig. 4A, lanes 3, 5). A similar expression pattern was found for the basic chitinases. After
Fig. 5. Expression of CBP20 in suspension-cultured cells after incubation with different heavy metals. Suspension-cultured cells were incubated overnight (15 h) without heavy metals (1), or with 100mM of each AgNO3(2), CdSO4(3), CoCl2(4), CuSO4(5) and ZnCl2(6), which were added to the cultures 6 days after subculturing. Subsequently the cells were extracted and used for immunoblotting.
Fig. 7. Influence of different heavy metals on the CBP20 mRNA (A) and protein level (B). Scratched leaves of in vitro plants were floated on water without heavy metals (1) and on 10 mM of each AgNO3(2), CdSO4(3), CoCl2(4), CuSO4(5) and ZnCl2(6).
100 mM ZnCl2 and without ZnCl2 and incubated
up to 7 days. At different time points, the cells were used for Northern blot experiments (Fig. 6). At the beginning of our experiment (up to the second day, lanes 1, 3) no CBP20 mRNA tran-scripts could be found. However, in nontreated controls the mRNA concentration increased later up to day 4 (lanes 5, 7) and decreased afterwards resulting in a slightly weaker signal at day 7 (lane 9). In the presence of 100 mM ZnCl2 (even lane
numbers), a very weak signal was obtained already after the first day of incubation (lane 2) which became more intensive with advancing incubation time in a similar manner as the control. The mRNA concentrations of the ZnCl2-treated cells
were higher up to day 4 (lane 8) than that of the control indicating an inducible effect of this heavy metal on the CBP20 expression.
Because suspension-cultured cells express CBP20 already without additional stress, we tested the effect of heavy metals on the expression of CBP20 in leaves. The same spectrum of heavy metals, which was tested on suspension cells, was applied to the fourth to the sixth leaf of in vitro plants. The leaves were scratched and floated for
15 h on distilled water containing 10 mM of the
different heavy metals and subsequently used for Western and Northern blotting experiments to register the CBP20 polypeptide and mRNA levels, respectively (Fig. 7A, B). No CBP20 protein was
detected in control leaves (Fig. 7B, lane 1) and in CoCl2-treated leaves (lane 4), a very weak signal
was found in AgNO3-treated leaves (lane 2) and a
rather stronger signal was seen in samples incu-bated with CuSO4(lane 5) and ZnCl2(lane 6). The
most intensive signal was obtained in CdSO4
-treated leaves (lane 3). In contrast, the highest CBP20 mRNA level was caused by ZnCl2 (Fig.
7A, lane 6) followed by CdSO4(lane 3) whereas no
mRNA transcript, could be detected in CoCl2- and
CuSO4- treated cells (lanes 4, 5). Either these two
heavy metals did not induce CBP20 gene expres-sion or they strongly impaired these cells. The missing mRNA in the CoCl2-treated culture could
be caused by toxic effects of CoCl2 at the used
concentration. This explains our failure to detect the CBP20 mRNA and protein. Surprisingly, our Western blotting experiment revealed two polypeptides of similar size which reacted with our CBP20 antibody. This was clearly seen in lanes 3, 5 and 6. A possible explanation could be that the heavy metals induce the expression of more than one gene of the small CBP20 gene family [10].
In our following experiments, the influence of different concentrations and incubation times of ZnCl2 were tested on leaves of in vitro plants.
Leaves of 4 – 6 weeks old plants were scratched and floated on water with 0, 0.1, 1, 10 and 100 mM ZnCl2 for 15 h. Subsequently, protein as
well as total RNA were extracted from parallel samples and used for detection of the CBP20 protein and mRNA, respectively (Fig. 8A,B). In all ZnCl2-treated leaves, CBP20 protein could be
detected. The most intensive polypeptide bands were found in samples treated with 10 and 100 mM ZnCl2 (Fig. 8B, lanes 4, 5). A definite higher
mRNA concentration was registered in leaves in-cubated with 0.1 mM ZnCl2 (Fig. 8A, lane 2)
compared to the control incubated without ZnCl2
Fig. 8. Influence of increasing ZnCl2 concentration on the CBP20 mRNA (A) and protein level (B). Young leaves of in vitro plants were dissected and floated on water without ZnCl2(1) and on water with 0.1 mM (2), 1 mM (3), 10 mM (4) and 100 mM (5) ZnCl2overnight.
reactions, and structural functions in nucleic acid metabolism, while the same heavy metals, and the even more potent ions of Cd, Hg, Ag, etc. are strongly poisonous to metal-sensitive enzymes, re-sulting in growth inhibition and death of the or-ganism. Heavy metals induce many changes in protein synthesis and gene expression [58,59].
It has also been shown that plants exposed to heavy metals respond by the accumulation of metal-binding polypeptides known as phy-tochelatins [60] and other stress-related proteins [61].
Chai et al. [62] described the expression of an 11 kDa proline-rich protein gene in bean in response to HgCl2, CdCl2, ZnCl2and CuSO4. Heavy metals
induce also glycine-rich proteins (GRPs) which are accumulated in the cell wall [61,63]. Interestingly, CBP20 also contains a glycin-rich region compris-ing 14 glycin residues and two additional G – G repeats. Recently, we described its extracellular accumulation in 2,4D-containing suspension cells [15] and it has been detected in preparations of the cell wall of these cultures as well (Hensel, unpub-lished), although CBP20 as basic representative of the PR-4 group was expected to be localized inside the cell, in the vacuole. However, in the case of the basic proteins chitinase and b-1,3-glucanase, we have made similar observations [14, unpublished]. We could speculate that special stress situations lead to an accumulation of CBP20 in the cell wall to protect these cells or neighbouring cells against damages caused by wounding, heavy metals and others by an unknown mechanism.
3.5. ZnCl2 does not change the stability of the CBP20 mRNA
An increasing CBP20 mRNA level can be caused either by a real induction realized by an increased synthesis of mRNA or by a changed stability of mRNA. To distinguish between both possibilities the mRNA synthesis registered after treatment with ZnCl2as well as in senescent leaves
was blocked by 100 mg ml−1 actinomycin D for 2
and 4 h. Nontreated leaves and leaves incubated with actinomycin D were used for Northern blot-ting experiments (Fig. 10A,B). Actinomycin D de-creased the CBP20 mRNA level. This reduction is stronger in ZnCl2-treated leaves (Fig. 10B) than in
old leaves (Fig. 10A). That means that the increase of the mRNA level caused by ZnCl2 is the result
(lane 1). No CBP20 mRNA transcripts were de-tected at higher ZnCl2 concentrations (1, 10, and
100 mM, lanes 3 – 5). Because the CBP20 protein was present at high concentrations it can be as-sumed that the CBP20 mRNA levels were tran-siently increased during the incubation and subsequently decreased up to the time point of analysis in consequence of the toxic compound. To test this suggestion shorter incubation periods each with 1 or 100 mM ZnCl2 were chosen to
follow the time course of CBP20 mRNA transcript levels (Fig. 9A,B). Thirty minutes after incubation in 1 mM ZnCl2, a strong signal could be registered
(Fig. 9A, lane 2) which decreased up to 4 h (lane 3) and increased again after 12 h of incubation (lane 4). The initial increase was very likely due to the stress caused by cutting off the leaves. How-ever, the second strong increase of mRNA concen-tration was induced by the metal ions. A total of 100 mM of ZnCl2 also caused an increased
con-centration of CBP20 mRNA, but this effect was only registered at 0.5 and 4 h (Fig. 9B, lanes 2, 3) of incubation and was lower compared with that of 1 mM ZnCl2.
Metal ions such as Cu2+ and Zn2+ are essential
trace nutrients taking part in redox reactions, elec-tron transfers, a multitude of enzyme catalyzed
Fig. 10. Analysis of the mRNA stability by blocking the mRNA synthesis with actinomycin D. Senescent leaves (A) and young leaves treated with 1 mM ZnCl2for 15 h to induce the CBP20 mRNA (B) were incubated without and with 100
mg ml−1actinomycin D for 2 and 4 h and were subsequently used for Northern blot analysis.
[4] D.J. Bowles, Defence-related proteins in higher plants, Annu. Rev. Biochem. 59 (1990) 873 – 907.
[5] H.J.M. Linthorst, Pathogenesis-related proteins of plants, Crit. Rev. Plant Sci. 10 (1991) 123 – 150. [6] R.F. White, J.F. Antoniw, Virus-induced resistance
re-sponses in plants, Crit. Rev. Plant Sci. 9 (1991) 443 – 455. [7] Y. Ohashi, M. Ohshima, Stress-induced expression of genes for pathogenesis-related proteins in plant, Plant Cell Physiol. 33 (1992) 819 – 826.
[8] J.F. Bol, A.S. Buchel, M. Knoester, T. Baladin, L.C. Van Loon, H.J.M. Linthorst, Regulation of the expres-sion of plant defence genes, Plant Growth Regul. 18 (1996) 87 – 91.
[9] A. Stintzi, T. Heitz, V. Prasad, S. Wiedemann-Merdi-noglu, S. Kauffmann, P. Geoffroy, et al., Plant ‘patho-genesis-related’ proteins and their role in defence against pathogens, Biochimie 75 (1993) 687 – 706.
[10] A.S. Ponstein, S.A. Bres-Vloemans, M.B. Sela-Buurlage, P.J.M. van den Elzen, L.S. Melchers, B.J.C. Cornelissen, A novel pathogen- and wound-inducible tobacco (Nico -tiana tabacum) protein with antifungal activity, Plant Physiol. 104 (1994) 109 – 118.
[11] T. Murashige, F. Skoog, A revised medium for rapid growth and bioassays with tobacco tissue cultures, Phys-iol. Plant 15 (1962) 473 – 497.
[12] R. Eichholz, J. Harper, G. Felix, F. Meins, Evidence for an abundant 33 000-Dalton polypeptide regulated by cy-tokinins in cultured tobacco tissues, Planta 158 (1983) 410 – 415.
[13] G. Felix, F. Meins, Ethylene regulation of b -1,3-glu-canase in tobacco, Planta 172 (1987) 386 – 392.
[14] I. Kunze, G. Kunze, M. Bro¨ker, R. Manteuffel, F. Meins, K. Mu¨ntz, Evidence for secretion of vacuolar
a-mannosidase, class I chitinase, and class I b -1,3-glu-canase in suspension cultures of tobacco cells, Planta 205 (1998) 92 – 99.
[15] G. Hensel, M. Brosius, I. Maeting, T. Wartmann, C. Horstmann, G. Kunze, et al., Cloning of the wound-in-ducible protein CBP20 and expression in suspension cultures of tobacco, Plant Sci. 128 (1997) 199 – 206. [16] J.J.M.R. Jacobs, K. Litie`re, V. van Dijk, G.J. van Eldik,
M. van Montagu, M. Cornelissen, Post-transcriptional
b-1,3-glucanase gene silencing involves increased tran-script turnover that is translation-independent, Plant J. 12 (1997) 885 – 893.
[17] J. Logemann, J. Schell, L. Willmitzer, Improved method for the isolation of RNA from plant tissues, Anal. Biochem. 163 (1987) 16 – 20.
[18] K. Herbers, G. Mo¨nke, R. Badur, U. Sonnewald, A simplified procedure for the subtractive cDNA cloning of photoassimilate-responding genes: isolation of cDNAs encoding a new class of pathogenesis-related proteins, Plant Mol. Biol. 29 (1995) 1027 – 1038.
[19] M.M. Bradford, A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding, Anal. Biochem. 72 (1976) 248 – 254.
[20] U.K. Laemmli, Cleavage of structural protein during assembly of the head of bacteriophage T 4, Nature (Lond.) 227 (1970) 680 – 685.
of a real induction and not of an increased mRNA stability of the CBP20 mRNA. But, a reduced turnover in case of aging cannot be excluded.
Exogenous signals have been found to affect mRNA stability of several plant genes [64 – 68]. Zhang et al. [68] have shown that treatment of cultured bean cells with a fungal elicitor decreased the transcript level without changing the appropri-ate transcription rappropri-ate.
Taken together the PR-protein CBP20 is an-other interesting stress protein with a similar be-haviour in its expression like other well-known basic PR-proteins but it differs from them in being inducible by SA. Therefore, CBP20 could be in-volved in a new signal transduction pathway.
Acknowledgements
The authors are grateful to H. Bohlmann and G. Oswald for skilful technical assistance. We also thank S. Sommer for critical reading of the manuscript. The experimental work was supported by the Kultusministerium des Landes Sachsen/ An-halt (859A/8284) as well as by the Deutsche Forschungsgemeinschaft (SFB363) and the Funds of Chemical Industry (Gotthard Kunze).
References
[1] L.C. Van Loon, Pathogenesis-related proteins, Plant Mol. Biol. 4 (1985) 111 – 116.
[2] J.P. Carr, D.F. Klessig, The pathogenesis-related proteins of plants, in: J.K. Setlow (Ed.), Genetic Engi-neering: Principles and Methods, vol. 11, Plenum Press New York, 1989, pp. 65 – 109.
[21] I. Kunze, G. Kunze, S. Ramm, C. Horstmann, R. Man-teuffel, K. Mu¨ntz, Two polypeptides that are secreted from suspension-cultured tobacco cells in the presence of brefeldin A have chitin-binding domains, J. Plant Phys-iol. 147 (1995) 63 – 70.
[22] C. Hanfrey, M. Fife, V. Buchanan-Wollaston, Leaf senescence in Brassica napus: expression of genes encod-ing pathogenesis-related proteins, Plant Mol. Biol. 30 (1996) 597 – 609.
[23] S. Potter, U. Scott, L. Kay, et al., Regulation of a hevein-like gene in Arabidopsis, Mol. Plant-Microbe In-teract. 6 (1993) 680 – 685.
[24] A. Stanford, M. Bevan, D. Northcote, Differential ex-pression within a family of novel wound-induced genes in potato, Mol. Gen. Genet. 215 (1989) 200 – 208.
[25] L. Friedrich, M. Moyer, E. Ward, J. Ryals, Pathogene-sis-related protein 4 is structurally homologous to the carboxy-terminal domains of hevein, Win1 and Win2, Mol. Gen. Genet. 230 (1991) 113 – 119.
[26] H.J.M. Linthorst, N. Danhash, F.T. Brederode, J.A.L. Van Kan, P.J.G.M. De Witt, J.F. Bol, Tobacco and tomato PR proteins homologous to win and pro-hevein lack the ‘hevein’ domain, Mol. Plant-Microbe Interact. 4 (1991) 585 – 592.
[27] W. Broekaert, H. Lee, A. Kush, N.H. Chua, N. Raikhel, Wound-induced accumulation of mRNA containing a hevein sequence in laticifers of rubber tree (He6ea
brasiliensis), Proc. Natl. Acad. Sci. USA 87 (1990) 7633 – 7637.
[28] B. Svensson, I. Svendsen, P. Hojrup, P. Roepstorff, S. Ludvigsen, F.M. Poulsen, Primary structure of barwin: a barley seed protein closely related to the C-terminal domain of proteins encoded by wound-induced plant genes, Biochemistry 31 (1992) 8767 – 8770.
[29] J. Hejgaard, S. Jacobsen, S.E. Bjorn, K.M. Kragh, Anti-fungal activity of chitin-binding PR-4 type proteins from barley grain and stressed leaf, FEBS Lett. 307 (1992) 389 – 392.
[30] C. Caruso, G. Chilosi, C. Caporale, L. Leonardi, L. Bertini, P. Magro, Induction of pathogenesis-related proteins in germinating wheat seeds infected withFusar -ium culmorum, Plant Sci. 140 (1999) 87 – 97.
[31] D.R. Lerner, N.V. Raikhel, The gene for stinging nettle lectin (Urtica dioicaagglutinin) encodes both a lectin and a chitinase, J. Biol. Chem. 267 (1992) 11085 – 11091. [32] J. Memelink, H.J. Linthorst, R.A. Schilperoort, J.H.
Hoge, Tobacco genes encoding acidic and basic isoforms of pathogenesis-related proteins display different expres-sion patterns, Plant Mol. Biol. 14 (1990) 119 – 126. [33] U.C. Biswal, B. Biswal, Ultrastructural modification and
biochemical changes during senescence of chloroplasts, Int. Rev. Cytol. 113 (1988) 271 – 321.
[34] L.D. Noode´n, Abscisic acid, auxin and other regulators of senescence, in: L.D. Noode´n, A.C. Leopold, (Eds.), Senescence and ageing in plants, Academic Press, San Diego, 1988, pp. 330 – 386.
[35] C.M. Smart, Gene expression during leaf senescence, New Phytol. 126 (1994) 419 – 448.
[36] H. Thomas, J.L. Stoddart, Leaf senescence, Annu. Rev. Plant Physiol. 31 (1980) 83 – 111.
[37] K.M. Davies, D. Grierson, Identification of cDNA clones for tomato (Lycopersicum esculentum Mill.) mR-NAs that accumulate during fruit ripening and leaf senescence in response to ethylene, Planta 179 (1989) 73 – 80.
[38] I. John, R. Hacket, W. Cooper, R. Drake, A. Farrel, D. Grierson, Cloning and characterization of tomato leaf senescence-related cDNAs, Plant Mol. Biol. 33 (1997) 641 – 651.
[39] V. Buchanan-Wollaston, C. Ainsworth, Leaf senescence inBrassica napus: cloning of senescence related genes by subtractive hybridisation, Plant Mol. Biol. 33 (1997) 821 – 834.
[40] V. Buchanan-Wollaston, Isolation of cDNA clones for genes that are expressed during leaf senescence inBras -sica napus, Plant Physiol. 105 (1994) 839 – 846.
[41] L.L. Hensel, V. Grbic, D.A. Baumgarten, A.B. Bleeker, Developmental and age-related processes that influence the longevity and senescence of photosynthetic tissues in Arabidopsis, Plant Cell 5 (1993) 553 – 564.
[42] J.I. Medford, J.S. Elmer, H.J. Klee, Molecular cloning and characterization of genes expressed in shoot apical meristems, Plant Cell 3 (1991) 359 – 370.
[43] W. Becker, K. Apel, Differences in gene expression be-tween natural and artificially induced leaf senescence, Planta 189 (1993) 74 – 79.
[44] Y. Azumi, A. Watanabe, Evidence for a senescence-asso-ciated gene induced by darkness, Plant Physiol. 95 (1991) 577 – 583.
[45] K.N. Lohman, S. Gran, M.C. John, R.M. Amasino, Molecular analysis of natural leaf senescence in Ara -bidopsis thaliana, Physiol. Plant 92 (1994) 322 – 328. [46] T. Lotan, N. Ori, R. Fluhr, Pathogenesis-related proteins
are developmentally regulated in tobacco flowers, Plant Cell 1 (1989) 881 – 887.
[47] R.S.S. Fraser, Evidence for the occurrence of the ‘patho-genesis-related’ proteins in leaves of healthy tobacco plants during flowering, Physiol. Plant Oath. 19 (1981) 69 – 76.
[48] F.Th. Brederode, H.J.M. Linthorst, J.F. Bol, Differential induction of acquired resistance and PR gene expression in tobacco by virus infection, ethephon treatment, UV light and wounding, Plant Mol. Biol. 17 (1991) 1117 – 1125.
[49] C.M.J. Pieterse, L.C. van Loon, Salicylic acid-indepen-dent plant defence pathways, TIPS 4 (1999) 52 – 58. [50] J. Hennig, R.E. Dewey, J.R. Cutt, D.F. Klessig,
Patho-gen, salicylic acid and developmental dependent expres-sion of ab-1,3-glucanase/GUS gene fusion in transgenic tobacco plants, Plant J. 4 (1993) 481 – 493.
[51] E.R. Ward, S.J. Uknes, S.C. Willias, S.S. Dincher, D.L. Wiederhold, D.C. Alexander, et al., Coordinate gene activity in response to agents that induce systemic ac-quired resistance, Plant Cell 3 (1991) 1085 – 1094. [52] N. Yalpani, P. Silverman, T.M.A. Wilson, D.A. Kleier,
I. Raskin, Salicylic acid is a systemic signal and an inducer of pathogenesis-related proteins in virus-infected tobacco, Plant Cell 3 (1991) 809 – 818.
ac-quired resistance but is reac-quired in signal transduction, Plant Cell 6 (1994) 959 – 965.
[54] Y. Ohashi, M. Matsuoka, Induction and secretion of pathogenesis-related proteins by salicylate or plant hor-mones in tobacco suspension cultures, Plant Cell Physiol. 28 (1987) 573 – 580.
[55] T. Niki, I. Mitsuhara, S. Seo, N. Ohtsubo, Y. Ohashi, Antagonistic effect of salicylic acid and jasmonic acid on the expression of pathogenesis-related (PR) protein genes in wounded mature tobacco leaves, Plant Cell Physiol. 39 (1998) 500 – 507.
[56] F. Meins, J.M. Neuhaus, C. Sperisen, J. Ryals, The primary structure of plant pathogenesis-related glu-canohydrolases and their genes, in: T. Boller, F. Meins (Eds.), Genes Involved in Plant Defence, Springer Ver-lag, 1991, pp. 245 – 282.
[57] S. Potter, S. Uknes, K. Lowton, A.M. Winter, D. Chan-dler, J. DiMaio, et al., Regulation of a hevein-like gene in Arabidopsis, Mol. Plant Microbe Interact. 6 (1993) 680 – 685.
[58] M. De Tapia, P. Bergman, A. Awade, G. Burkard, Analysis of acid extractable bean leaf proteins induced by mercuric chloride treatment and alfalfa mosaic virus infection. Partial purification and characterization, Plant Sci. 45 (1986) 167 – 177.
[59] M. Margis-Pinheiro, C. Martin, L. Didierjan, G. Burkard, Differential expression of bean chitinase genes by virus infection, chemical treatment and UV irradia-tion, Plant Mol. Biol. 22 (1993) 659 – 668.
[60] J.C. Steffens, The heavy metal-binding peptides of plants, Annu. Rev. Plant Physiol. Plant Mol. Biol. 41 (1990) 553 – 575.
[61] L. Didierjan, P. Frendo, W. Nasser, G. Genot, J. Marivet, G. Burkard, Heavy-metal-responsive genes in maize: identification and comparison of their expression upon various forms of abiotic stress, Planta 199 (1996) 1 – 8.
[62] T.Y. Chai, L. Didierjan, G. Burkard, G. Genot, Expres-sion of a green tissue-specific 11 kDa proline-rich protein gene in bean in response to heavy metals, Plant Sci. 133 (1998) 47 – 56.
[63] L. Didierjan, P. Frendo, G. Burkard, Stress response in maize: sequence analysis of cDNAs encoding glycine-rich proteins, Plant Mol. Biol. 18 (1992) 847 – 849.
[64] L.F. Dickey, M. Gallo-Meagher, W.F. Thompson, Light regulatory sequences are located within the 5%portion of
theFed-1 message sequence, EMBO J. 11 (1992) 2311 – 2317.
[65] Y. Li, T.J. Strabala, G. Hagen, T.J. Guilfoyle, The soybean SAUR open reading frame contains a cis ele-ment responsible for cycloheximide-induced mRNA ac-cumulation, Plant Mol. Biol. 24 (1994) 715 – 723. [66] J.E. Lincoln, R.L. Fisher, Diverse mechanisms for the
regulation of ethylene-inducible gene expression, Mol. Gen. Genet. 212 (1988) 71 – 75.
[67] B.W. Shirley, R.B. Meagher, A potential role for RNA turnover in the light regulation of plant gene expression: ribulose-1,5-biphosphate carboxylase small subunit in soybean, Nucl. Acids Res. 18 (1990) 3377 – 3385. [68] S. Zhang, J. Sheng, Y. Liu, M.C. Mehdy, Fungal
elici-tor-induced bean proline-rich protein mRNA down-regu-lation is due to destabilization that is transcription and translation dependent, Plant Cell 5 (1993) 1089 – 1099.