Cardiol Young 2009: Page 1 of 4
rCambridge University Press ISSN 1047-9511 doi:10.1017/S1047951109990813
Original Article
Genetic screening of 104 patients with congenitally malformed
hearts revealed a fresh mutation of
GATA4
in those with atrial
septal defects
Haruka Hamanoue,1,2 Sri Endah Rahayuningsih,4 Yuya Hirahara,1 Junko Itoh,1 Utako Yokoyama,3 Takeshi Mizuguchi,1 Hirotomo Saitsu,1 Noriko Miyake,1 Fumiki Hirahara,2 Naomichi Matsumoto1 1
Department of Human Genetics; 2Department of Obstetrics and Gynecology;3Cardiovascular Research Institute, Yokohama City University Graduate School of Medicine, Yokohama, Japan; 4Department of Pediatrics, Padjadjaran University Medical School, Hasan Sadikin General Hospital, Bandung, Indonesia
Abstract We analysed the GATA binding protein 4 gene, or GATA4, along with the NK2 transcription factor related, locus 5 gene, orNKX2.5, to determine their genetic contribution to 104 sporadic patients in Indonesia with congenitally malformed hearts, 76 cases having atrial septal defect and 28 tetralogy of Fallot. We found only 1 novel mutation of GATA4 in those with atrial septal defecst. Analysis of the genetic background of the parents of the patient showed for the first time that a new mutation of GATA4can cause sporadic atrial septal defects. We failed to discover any other mutations of either the GATA4 or NKX2-5
genes, supporting the marked genetic heterogeneity of human congenital cardiac defects.
Keywords: genetic inheritance; new mutation; interatrial communications
C
ONGENITAL CARDIAC DISEASE IS ONE OF THE commonest defects found at birth, being observed in more than 1 of each 100 live births.1 Among these lesions, atrial septal defect and tetralogy of Fallot account for approximately one-tenth and one-twentieth of the overall defects, respectively. Genetic factors have been associated with both lesions. Mutations ofGATA4were found mostly in familial examples of atrial septal defect,2–9 but never in these lesions when occurring sporadically. Mutations of NKX2-5 were also observed in a spectrum of both familial and sporadic occurrences of congenitally malformed hearts, including those with atrial septal defect and tetralogy of Fallot, frequently in association with abnormalities of atrioventricular conduction.10–17In this study, we sequencedGATA4andNKX2-5
to determine their genetic contribution to the occurrence of sporadic congenital cardiac defects in Indonesia, producing what is, to the best of our knowledge, the first evidence of a new mutation of
GATA4causing an atrial septal defect.
Materials and methods
We recruited 76 patients with atrial septal defects located within the oval fossa, and 28 patients with tetralogy of Fallot. In Table 1, we list the cardiac phenotypes. All patients were non-syndromic, and had been diagnosed on the basis of echocardiogra-phy and other standard methods by one of coauthors (S.E.R.) working at Hasan Sadikin General Hospital in Bandung, Indonesia. As established by taking a careful history, we confirmed that none of the parents possessed an abnormal cardiac phenotype. Informed consent was obtained from all the participating families. As normal controls were unavailable for the Indonesian population, we used
Correspondence to: Naomichi Matsumoto, M.D., Ph.D., Department of Human Genetics, Yokohama City University Graduate School of Medicine, Fukuura 3-9, Kanazawa-ku, Yokohama 236-0004, Japan. Tel:181-45-787-2604; Fax:
normal Japanese controls to observe polymorph-isms, recognizing that Indonesian and Japanese populations are ethnically unrelated. Experimental protocols were approved by the institutional review board in Yokohama city university school of medicine.
Genomic deoxyribonucleic acid, or DNA, was extracted from peripheral leukocytes using FlexiGene Blood DNA kit (Qiagen, Tokyo, Japan). Whole genome amplification was performed wtih Illustra GenomiPhi V2 DNA Amplification Kit (GE Healthcare Bio-Science, Tokyo, Japan).
We analysed 6 coding exons of GATA4, and 2 exons ofNKX2-5, along with their flanking intronic regions. All primers were designed using the web-based Primer3 software (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi). Detailed information of primers is available on request. Polymerase chain
reaction was cycled 35 times at 948C for 30 sec, at 58–648C for 30 sec and at 728C for 30 sec in a total volume of 20ml containing 30 ng of whole genome amplification deoxyribonucleic acid, and/or genomic deoxyribonucleic acid, as a template, 0.5mM of forward and reverse primers, 200mM of each deoxynucleotide triphosphates, and 0.5 Unit of ExTaq (Takara, Ohtsu, Japan) or LATaq (Takara, Japan), purified with ExoSAP-IT (United States Biochemical, Cleveland, OH), and sequenced using BigDye Terminator ver.3.1 (Applied Biosystems, Foster City, CA) on the 3100 Genetic Analyzer (Applied Biosystems). Deoxyribonucleic acid sequences were compared with the reference genome sequences based on the University of California, Santa Cruz Genome Browser (May 2006 assembly) using the SeqScape software Ver.2.1 (Applied Biosystems). All the nucleotide changes found in whole genome amplifica-tion deoxyribonucleic acids were also confirmed by sequencing using original genomic deoxyribonucleic acids. Regarding polymorphisms, normal control deoxyribonucleic acids were sequenced until the change was seen, needing up to 96 normal chromosomes.
Results and discussion
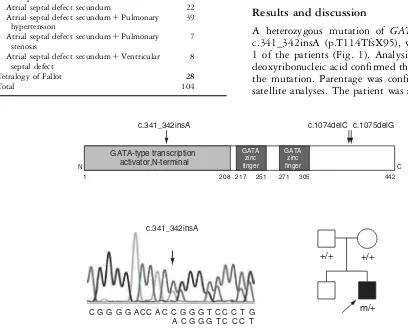
A heterozygous mutation of GATA4, specifically c.341_342insA (p.T114TfsX95), was identified in 1 of the patients (Fig. 1). Analysis of his parental deoxyribonucleic acid confirmed the novel nature of the mutation. Parentage was confirmed by micro-satellite analyses. The patient was a 4-year-old boy, Table 1. Cardiac phenotypes of the patients.
Cardiac phenotype
Number of patients
Atrial septal defect 76
Atrial septal defect secundum 22 Atrial septal defect secundum1Pulmonary
hypertension
39 Atrial septal defect secundum1Pulmonary
stenosis
7 Atrial septal defect secundum1Ventricular
septal defect
8
Tetralogy of Fallot 28
Total 104
The protein truncation mutation of GATA4. c.341_342insA identified in this study is the earliest type of truncation. c.1074delC and c.1075delG were previously reported (upper).2,3Electropherogram of the identified mutation is shown (lower left). The family pedigree of the patient with c.341_342ins shows clearly its new occurrence.1: wild type, m: mutated.
presenting because of failure to thrive and recurrent respiratory infections. Echocardiography revealed a hole of 1.4 centimeters diameter in the floor of the oval fossa, as well as pulmonary hypertension. At the age of 4 1/2 years, the defect was closed by insertion of an Amplatzer septal occluder. He is now in a good condition. His healthy 30-year-old father, and 20-year-old mother, were not consanguineous. The pregnancy had been uneventful, and his elder brother, aged 6 years, is also healthy, without any cardiac or other abnormalities (Fig. 1).
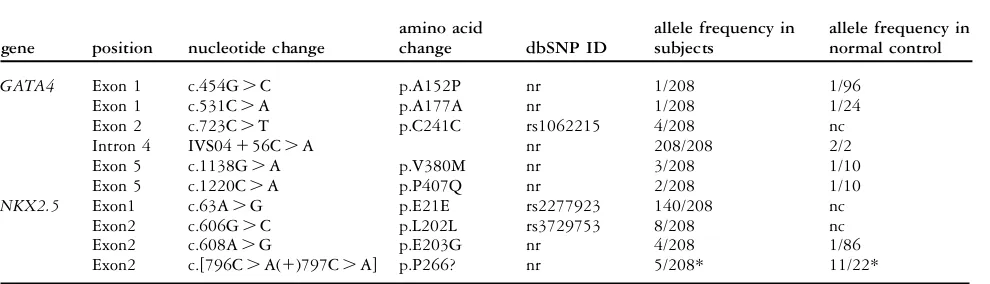
The mutation, c.341_342insA, is the third protein truncation mutation of GATA4, resulting in much earlier termination than the other two, c.1074delC and c.1075delG, thus far reported.2,3,5 All the reported truncations have been found in patients with atrial septal defects. Missense muta-tions are associated with varied phenotypes, includ-ing atrial septal defect, tetralogy of Fallot, atrioventricular septal defect, and ventricular septal defect.2,4–7,9,18. Three inframe changes, specifically p.S46del, p.A118_A119insA, and p.A125_A126in sAA, have also been found in patients with ventricular septal defect and tetralogy of Fallot.18 Mutations ofGATA4mutations have been reported to be very rare in patients with sporadic congenital cardiac lesions, with an estimated prevalence of less than 1%,8,9,18–20 this fitting well with our own finding. We also found 5 other SNPs, including rs1062245 inGATA4, as judged by normal control studies or according to the database of single nucleotide polymorphisms (http://www.ncbi.nlm. nih.gov/sites/entrez) (Table 2). GATA4, a central zinc finger transcription factor for cardiac develop-ment, binds specifically to the A/T GATA A/G motif on the deoxyribonucleic acid, and modulates transcription of downstream target genes.9 GATA4 can also physically interact with NKX2-521 and TBX5,2 producing familial congenital heart de-fects12 as well as the well recognized Holt-Oram
syndrome, which consists of cardiac septal defects, abnormalities of conduction, and skeletal anomalies of the limbs.22,23The previously reported mutation, c.1075delG, failed to activate transcription of downstream genes.2 Thus mutational consequences of the c.341_342insA found in this study could be similar to c.1075delG as c.341_342insA, and may potentially produce a much shorter protein than c.1075delG, even if its transcript escapes the nonsense mediated decay (Fig. 1).
None of our patients presented with any pathological changes inNKX2.5, albeit that 5 polymorphisms have been identified in this gene (Table 2). Approximately one-twentieth of examples of both atrial septal defect and tetralogy of Fallot may be caused by such mutations.11 The fact that none of our patients exhibited such abnormalities is further evidence supporting the notion that sporadic examples of congenital cardiac disease are genetically heterogeneous. Our analysis of 104 patients with sporadic forms of atrial septal defect and tetralogy of Fallot, therefore, for mutations of the GATA4 and NKX2-5 genes has revealed a new mutation ofGATA4 in 1 patient with an atrial septal defect. As far as we are aware, this is the first direct link of such a new mutation demonstrated using genomic deoxyribonucleic acids.
Acknowledgments
We express our gratitude to all patients and their families who participated in this project. This study was supported by a Research Grant from the Ministry of Health, Labour and Welfare (N. M.).
Contract grant sponsor: Research Grant from the Ministry of Health, Labour and Welfare (N. M.).
References
1. Hoffman JI, Kaplan S. The incidence of congenital heart disease. J Am Coll Cardiol 2002; 39: 1890–1900.
Table 2. Polymorphisms found in this study.
gene position nucleotide change
amino acid
change dbSNP ID
allele frequency in subjects
allele frequency in normal control
GATA4 Exon 1 c.454G.C p.A152P nr 1/208 1/96
Exon 1 c.531C.A p.A177A nr 1/208 1/24
Exon 2 c.723C.T p.C241C rs1062215 4/208 nc
Intron 4 IVS04156C.A nr 208/208 2/2
Exon 5 c.1138G.A p.V380M nr 3/208 1/10
Exon 5 c.1220C.A p.P407Q nr 2/208 1/10
NKX2.5 Exon1 c.63A.G p.E21E rs2277923 140/208 nc
Exon2 c.606G.C p.L202L rs3729753 8/208 nc
Exon2 c.608A.G p.E203G nr 4/208 1/86
Exon2 c.[796C.A(1)797C.A] p.P266? nr 5/208* 11/22*
Normal controls are Japanese; nr: not registered; nc: not checked; *: if both changes are in the same allle.
2. Garg V, Kathiriya IS, Barnes R, et al. GATA4 mutations cause human congenital heart defects and reveal an interaction with TBX5. Nature 2003; 424: 443–447.
3. Okubo A, Miyoshi O, Baba K, et al. A novel GATA4 mutation completely segregated with atrial septal defect in a large Japanese family. J Med Genet 2004; 41: e97.
4. Sarkozy A, Conti E, Neri C, et al. Spectrum of atrial septal defects associated with mutations of NKX2.5 and GATA4 transcription factors. J Med Genet 2005; 42: e16.
5. Hirayama-Yamada K, Kamisago M, Akimoto K, et al. Pheno-types with GATA4 or NKX2.5 mutations in familial atrial septal defect. Am J Med Genet A 2005; 135: 47–52.
6. Tomita-Mitchell A, Maslen CL, Morris CD, Garg V, Goldmuntz E. GATA4 sequence variants in patients with congenital heart disease. J Med Genet 2007; 44: 779–783.
7. Rajagopal SK, Ma Q, Obler D, et al. Spectrum of heart disease associated with murine and human GATA4 mutation. J Mol Cell Cardiol 2007; 43: 677–685.
8. Posch MG, Perrot A, Schmitt K, et al. Mutations in GATA4, NKX2.5, CRELD1, and BMP4 are infrequently found in patients with congenital cardiac septal defects. Am J Med Genet A 2008; 146: 251–253.
9. Nemer G, Fadlalah F, Usta J, et al. A novel mutation in the GATA4 gene in patients with Tetralogy of Fallot. Hum Mutat 2006; 27: 293–294.
10. Akazawa H, Komuro I. Cardiac transcription factor Csx/Nkx2-5: Its role in cardiac development and diseases. Pharmacol Ther 2005; 107: 252–268.
11. McElhinney DB, Geiger E, Blinder J, Benson DW, Goldmuntz E. NKX2.5 mutations in patients with congenital heart disease. J Am Coll Cardiol 2003; 42: 1650–1655.
12. Schott JJ, Benson DW, Basson CT, et al. Congenital heart disease caused by mutations in the transcription factor NKX2-5. Science 1998; 281: 108–111.
13. Benson DW, Silberbach GM, Kavanaugh-McHugh A, et al. Mutations in the cardiac transcription factor NKX2.5 affect
diverse cardiac developmental pathways. J Clin Invest 1999; 104: 1567–1573.
14. Goldmuntz E, Geiger E, Benson DW. NKX2.5 mutations in patients with tetralogy of fallot. Circulation 2001; 104: 2565–2568.
15. Reamon-Buettner SM, Hecker H, Spanel-Borowski K, et al. Novel NKX2-5 mutations in diseased heart tissues of patients with cardiac malformations. Am J Pathol 2004; 164: 2117–2125.
16. Watanabe Y, Benson DW, Yano S, et al. Two novel frameshift mutations in NKX2.5 result in novel features including visceral inversus and sinus venosus type ASD. J Med Genet 2002; 39: 807–811.
17. Ikeda Y, Hiroi Y, Hosoda T, et al. Novel point mutation in the cardiac transcription factor CSX/NKX2.5 associated with con-genital heart disease. Circ J 2002; 66: 561–563.
18. Zhang W, Li X, Shen A, et al. GATA4 mutations in 486 Chinese patients with congenital heart disease. Eur J Med Genet 2008; 51: 527–535.
19. Schluterman MK, Krysiak AE, Kathiriya IS, et al. Screening and biochemical analysis of GATA4 sequence variations identified in patients with congenital heart disease. Am J Med Genet A 2007; 143: 817–823.
20. Posch MG, Berger F, Perrot A, Ozcelik C. We need a detailed phenome in the phenomenon of genetics and congenital heart disease. J Med Genet 2008; 45: 320.
21. Zhu W, Shiojima I, Hiroi Y, et al. Functional analyses of three Csx/Nkx-2.5 mutations that cause human congenital heart disease. J Biol Chem 2000; 275: 35291–35296.
22. Basson CT, Bachinsky DR, Lin RC, et al. Mutations in human TBX5[corrected]cause limb and cardiac malformation in Holt-Oram syndrome. Nat Genet 1997; 15: 30–35.
[23. Li QY, Newbury-Ecob RA, Terrett JA, et al. Holt-Oram syndrome is caused by mutations in TBX5, a member of the Brachyury (T) gene family. Nat Genet 1997; 15: 21–29.