Structure of a glutathione peroxidase homologous gene involved in
the oxidative stress response in
Chlamydomonas reinhardtii
Urs Leisinger, Karin Ru¨fenacht, Alexander J.B. Zehnder, Rik I.L. Eggen *
Department of Microbiology,Swiss Federal Institute for En6ironmental Science and Technology(EAWAG),Ueberlandstrasse133,
CH-8600Duebendorf,Switzerland
Received 25 February 1999; received in revised form 26 July 1999; accepted 26 July 1999
Abstract
The organisation and nucleotide sequence of the single copy glutathione peroxidase homologous gene gpxh from Chlamy
-domonas reinhardtiiis reported. Thegpxhgene consists of five exons and four introns, and encodes a predicted protein (GPXH) of 162 amino acids. GPXH belongs to the family of glutathione peroxidase (GPX)-like proteins and showed high homology with the deduced amino acid sequences ofgpx-related genes from yeast (67 – 78% similarity) and from plants (60 – 65% similarity). The GPXH from C.reinhardtii differs from the well characterized mammalian cytosolic GPX (GPX1) in that it contains a normal cysteine residue instead of a selenocysteine, that the residues responsible for glutathione binding at the reactive center in GPX1 are not present, and that two amino acid stretches important for the tetramerisation of GPX1 are absent. Northern blot experiments revealed a single 1.3 kb mRNA of which the cellular concentration is elevated strongly upon exposure to chemicals causing oxidative stress. In addition, salt stress did cause a weak increase in mRNA concentration. This indicates thatgpxhis an oxidative stress responding gene rather than a general stress responsive gene. © 1999 Elsevier Science Ireland Ltd. All rights reserved.
Keywords:Hydrogen peroxide; Organic hydroperoxide; Paraquat; Transcriptional regulation
www.elsevier.com/locate/plantsci
1. Introduction
Glutathione peroxidase (GPX) catalyses the re-duction of hydrogen peroxide (H2O2) or organic hydroperoxides to water or alcohols by reduced glutathione (GSH). Since the discovery of GPX in 1957 [1], it has been a thoroughly investigated protein shown to be important in the defense against oxidative stress [2]. The initially character-ized mammalian cytosolic GPX (GPX1) exists as a homotetramer with a subunit mass of 21 kDa [3] and contains a selenocysteine (SeCys) residue at the active site. Later, several additional types of mammalian GPX were isolated, including the Se-independent epididymal secretory GPX (GPX5) that was first isolated from mice [4].
GPX-like activities have not only been mea-sured in mammals, but also in yeast, in algae and in higher plants [5]. In some of these cases the enzymes exhibiting the GPX-like activities might
have been glutathione S-transferases (GST) that
are known to exhibit GPX-activity besides their GST activity. In other cases, GPX rather than GST were found: a GPX with an activity restricted to the reduction of organic peroxides has been isolated from the yeastHansenula mrakii [6], and a GPX that metabolized at least some organic
per-oxides as well as H2O2 was found in
Chlamy-domonas reinhardtii [7]. The latter has been shown to contain selenium and appears to be of te-trameric structure, like the mammalian GPX1. No other plant GPX-like protein has so far been shown to contain selenium or to have a tetrameric nature.
* Corresponding author. Tel.: +41-1-8235320; fax: + 41-1-8235547.
E-mail address:[email protected] (R.I.L. Eggen)
In order to examine the structure, the function, and the regulation of the enzymes with GPX-like activities, attempts were made to isolate the corre-sponding genes. Indeed, several gpx-related genes have recently been isolated from bacteria [8,9],
from the nematode Brugia pahangi [10], from the
helminth Schistosoma mansonii [11], from
Plas-modium falciparum [12], from yeast (unpublished, accession numbers P38143, P40581, 36014 (swis-sprot) AB012395 (EMBL)) and from higher plants [5,13 – 15]. In contrast to the mammalian GPX1
but like GPX5, all these non vertebrate
gpx-re-lated genes, except for the SeCys-encoding gene isolated fromS. mansonii, carry a codon for a Cys residue at the putative catalytic site instead of the TGA codon for SeCys. This is a striking difference to GPX1, because selenium atoms are essential for catalysis of GPX1 and the replacement of sele-nium by sulfur results in a reduction of protein activity by more than 99%. This can be explained by the fact that selenium leads to more nucle-ophilic power and a lower pK than sulfur. There-fore it reacts much faster with hydroperoxides [16]. For the Cys-(instead of SeCys) encoding gpx-like genes it was unclear, whether they encoded for functional GPXs and what their physiological role was. Further studies have revealed that the Cys-containing GPX-like proteins from Citrus sinensis and B. pahangi exhibit GPX activity towards or-ganic hydroperoxides [17,18], even though this activity is considerably lower than reported for the SeCys-containing GPXs. Thus, alternative roles for Cys-containing GPX-like proteins may exist in vivo. Further functional studies were consistent with an important role of selenium independent GPX-like proteins in the cellular stress response: The GPX-like proteins from N. syl6estris and C.
sinensis showed elevated expression under Hg-stress and salt Hg-stress respectively [19,20], Es -cherichia colicells that express thegpx-like gene of the Citrus plant are more resistant against oxida-tive stress excerted by paraquat [21], and clones of Neisseria meningitidis with an interrupted gpx-like gene show higher sensitivity towards paraquat [22]. The mammalian Se-independent GPX5 has been shown to have GPX activity and to protect cells against elevated concentrations of H2O2 in the culture medium [23]. In this report, we de-scribe the isolation and characterization of a
gpx-related gene from C. reinhardtii. Since C.
reinhardtiiis accessible to a wide variety of genetic
and molecular techniques [24,25], it appeared well suited for studying the physiological role of this gene. The work presented here is a first step in our investigation of the oxidative stress response in C. reinhardtiiand shows that gpxhplays a role in the response to oxidative stress.
2. Methods
2.1. Strains and culture conditions
C. reinhardtii strain cw15arg7mt- (CC-325) was cultured in Sueoka high salt medium HSM or Tris – acetate – phosphate-medium TAP [26]. The cultures were agitated on rotatory shakers (150 rpm) under constant illumination of 40 mmol m−2
s−1 photosynthetically active radiation at 25°C (standard conditions). Cells were grown on soli-dified HSM containing 1% agar under a cycle of 14 h illumination (25mmol m−2s−1
photosynthet-ically active radiation) and 10 h dark at 25°C. Media were supplemented with 50 mg/l arginine, 50 mg/l ampicilline and 1.2 g/l sodium acetate when required.
E. coli DH5a [27] was used for routine cloning
experiments and was grown on LB at 37°C.E. coli BL21(DE3)(pLysS) [28] was used for the expres-sion of pET8c derived plasmids [29].
2.2. Nucleic acid preparation
Genomic DNA of exponentially growing cul-tures of C. reinhardtiicw15arg7mt- was isolated by lysing cells in 100 mM Tris – HCl pH 8.0, 2% CTAB, 1.4 M NaCl, 20 mM EDTA and 2% b-Mercaptoethanol at 65°C for 2 h, extracting the lysate with phenol, precipitating contaminating polysaccharides in 1% CTAB, 70 mM NaCl, 1 vol. chloroform/isoamyl alcohol, and by a final precip-itation of the DNA with isopropanol [30].
Total RNA was prepared by the acid guanidine isothiocyanate – phenol – chloroform method [31] using TRIzol Reagent (Life Technologies Ltd.).
2.3. Construction of cDNA library
Three 100-ml TAP cultures of C. reinhardtii
respectively and grown for an additional 4 h. The three cultures were pooled and poly(A)-containing RNA was isolated using the fast track mRNA isolation kit (Invitrogen).
Custom cDNA library construction and direc-tional cloning into the yeast expression vector pYES2 has been done by Invitrogen. The library consists of 106 primary recombinants with a mean insert size of 1 kb.
2.4. Isolation of full length cDNA and genomic DNA of gpxh
Total RNA fromC. reinhardtiicw15arg7mt- was
employed in a reverse transcriptase reaction (RT) using the ready-to-go T-primed first strand synthe-sis kit (Pharmacia) according to the manufactur-er’s directions. The single stranded cDNA served as a template for PCR with a pre-cycle incuba-tion at 94°C for 2 min., and subsequent 35 cycles of 1 min at 94°C, 1 min at 60°C, and 90 s at 72°, followed by a final incubation at 72°C for
4 min. As primers, the RT-primer (5%
-AACTGGAAGAATTCGCGGCCGCAGGAAT18
-3%) and a degenerated oligonucleotide (5%
-GCMT-TYCCBTGYAAYCARTTYGG-3%) derived from
a consensus amino acid sequence (AFPCNQFG) in glutathione peroxidases were used. The PCR product was cloned via TA cloning into the pT7blue vector (Novagen) and used as a probe for
the isolation of gpxh containing cDNA and
ge-nomic DNA from libraries using the colony blot-ting protocol of Ausubel et al. [30]. For stringent
hybridizations, the final wash in 0.1×SSC and
0.1% SDS was carried out at 63°C. For non stringent hybridizations, the final wash was carried
out in 2×SSC and 0.1% SDS at 60°C. At washing
temperatures below 60°C, a strong unspecific background was obtained. A first screening of 500 000 clones of the cDNA library revealed 20 positive plaques. In order to find cDNA clones with a potential complete 5% end, PCR
amplifica-tion was conducted on positive plaques of the cDNA library with a primer complementary to the multiple cloning site in pYES2 vector upstream from the 5% end of the cloned cDNA insert (5%
-CGACTCACTATAGGGAATATTAAGC-3%)
and a primer complementary to the coding strand of the cDNA as derived from the RT-PCR
product (5%
-AATGGGGAACGTAACGCCGAA-3%). PCR amplification conditions were: pre-cycle
incubation at 94°C for 2 min, and subsequent 35 cycles of 50 s at 94°C, 1 min at 60°C and 2 min at 72°C, followed by a final incubation at 72°C for 4 min. The colonies from the area of the plaque that revealed the longest PCR product were removed and plated out for a second screening. After a third analogous screening step, a single positive colony, designated pGPXa3, could be isolated and
was chosen for further analysis.
In order to obtain genomic DNA containing the
gpxh gene, 50 000 clones of a cosmid library,
which was kindly, provided by Donald. P. Weeks [32] were screened. Cosmid cGPX1.4 was selected
out of six positive colonies for further
investigation.
2.5. O6erexpression of gpxh in E. coli
DNA fragments containing the gpxh coding
se-quence were cloned into pET8c [29] and
intro-duced into E. coli BL21(DE3)(pLysS) [28]. The
transformants were grown at 25°C in LB medium
to an OD600=0.5, overexpression was induced
with 1 mM IPTG and cells grown to OD600=1.0.
Crude extract was prepared in the presence of 1 mg/l pefabloc sc proteinase inhibitor (Boehringer) using a french press.
2.6. Dynamics of gpxh mRNA
Cultures of C. reinhardtii cw15arg7mt- were grown to a density of 1 – 4×106 cells per ml in
HSM, exposed to 0.2 mM H2O2, 0.2 mM
tert-butyl hydroperoxide (t-BOOH), 0.5 mM Paraquat
or 50 – 200 mM NaCl and total RNA was ex-tracted from aliquots taken at regular intervals. Aliquots of 20mg of total RNA were denatured
with glyoxal/DMSO, blotted, hybridized and
washed following the protocol of Ausubel et al.
[30]. The last wash in 0.1×SSC and 0.1% SDS
was carried out at 63°C. The amount of total RNA was controlled by staining with ethidium bromide.
2.7. Sequencing
Fragments of cosmid clone cGPX1.4 and cDNA clone pGPXa3 were subcloned. The fragments of
pGPXa3 were sequenced in both directions
DNA were sequenced only in one direction. Se-quencing reactions with IRD41-labelled primers M13universal and M13reverse (MWG-Biotech) were carried out with the labstation thermo seque-nase labeled primer cycle sequencing kit (Amer-sham) following the manufacturer’s instructions and they were analyzed on a LI-COR sequencer 4000 using the Base ImagIR software package (LI-COR). Accession number of the sequence on EMBL and GenBank: AF014927.
2.8. Primer extension
Primer extension was carried out in an opti-mized reaction mixture with two different IRD41
labeled primers (IRGPX5.1: 5%
-GGTGTTAGGC-GATTGGTTATTG-3%; IRGPX5.4: 5%
-TCGTCGACAGGCCGTAAAACT-3%) (MWG
biotech) complementary to the 5% untranslated
re-gion of the gpxh mRNA. In a total volume of 14
ml, 20 mg of total RNA, 10 pmol of one primer, 1
ml DMSO and water were incubated at 93°C for 2
min and directly cooled to 42°C. Subsequently, 4
ml of a 5× AMV RT buffer (Amersham), 1 ml
dNTP mix (10 mM each) and 0.5 ml AMV RT (5
U) were added and the reaction was performed for 1 h at 42°C. After heating the reaction mixture to 95°C for 5 min, the RNA was digested with 10 ng RNAse at 37°C for 30 min. The cDNA was pre-cipitated with ethanol and analyzed on a LI-COR sequencer 4000.
To determine the size of the cDNA, the same primers were applied in a sequencing reaction on the corresponding chromosomal DNA.
3. Results
3.1. Isolation of gpxh genomic DNA and cDNA
A glutathione peroxidase homologous gene
(gpxh) has been isolated from C. reinhardtii by
reverse genetics. Total RNA extracted from C.
reinhardtii was used to generate single stranded
cDNA with oligo dT18 as a primer. This cDNA
served as a template in a PCR using the dT18
primer in combination with a degenerate primer deduced from conserved amino acid sequences in GPX-like proteins from other organisms. The se-quence of the obtained band contained an open reading frame with a deduced amino acid sequence
exhibiting high homology to known glutathione peroxidases.
This PCR product was used as a probe to screen
a cDNA and a cosmid library of C. reinhardtii.
Out of 500 000 screened cDNA clones, 20 clones hybridized with the probe. From these, the cDNA clone pGPXa3 with the longest 5%region as
deter-mined by PCR analysis was used for further stud-ies. Among 50 000 cosmid clones, six clones hybridized with the PCR product. In a southern blot analysis, the clone named cGPX1.4 proved to
contain the longest gpxh-containing chromosomal
fragment and thus was chosen for subcloning and further analysis.
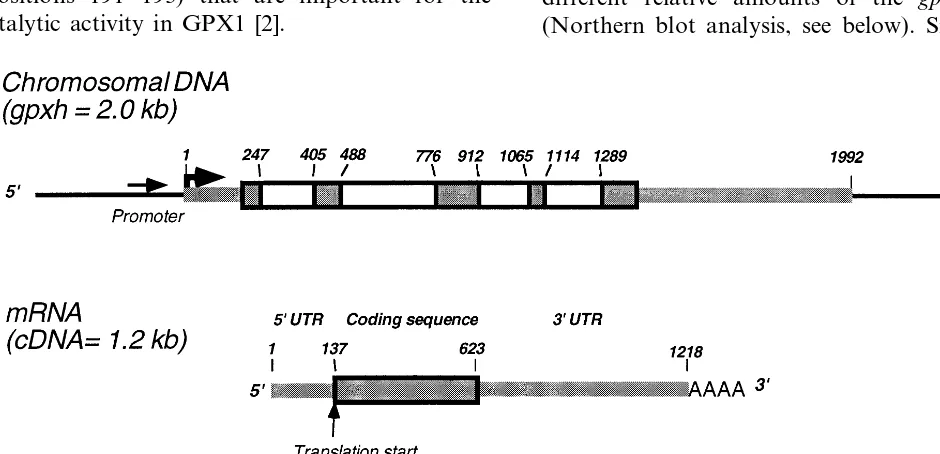
3.2. Gene organization
Southern blot analysis, performed under low stringency conditions, with the full-length cDNA clone as a probe revealed that the C. reinhardtii gpxh is a single copy gene (data not shown). This result was supported by hybridization patterns of cDNA and cosmid libraries under stringent and non-stringent washing conditions. The stringency of the washings did not influence the hybridization pattern. In view of the fact that Yokota et al. [7]
have isolated a Se-dependent GPX from C.
rein-hardtii, which is related to mammalian GPX1, this is an unexplained, remarkable result. Sequence analysis and primer extension experiments indi-cated that the dominant (see below) 1218
nucle-otides long gpxh mRNA contained a 5%
untranslated region of 137 nucleotides followed by an open reading frame of 486 nucleotides and a 3%
untranslated region of 592 nucleotides. The puta-tive translation start codon is flanked by a purine
in position −3 and G in position +4
(A6 GCATGG) characteristic for strong eucaryotic
initiator codons [33]. The 3% end of the mRNA
carries a putative polyadenylation signal
(TG-TAA) found in other Chlamydomonas genes
[26,34] and ends in a poly(A)-tail. Comparison of the cDNA with genomic DNA sequences implied
that the gpxh gene is composed of five exons as
shown schematically in Fig. 1. The four introns were localized in the open reading frame region of
gpxh and their boundaries were verified by the
eucaryotic splice junction consensus sequence [35].
As observed for other C. reinhardtii genes, the
introns are relatively small ranging from 153 to
288 bp. The GC content of the gpxh mRNA is
other C. reinhardtii nuclear genes of 64.0% [36].
The codon usage of the gpxh reflects the codon
usage of C. reinhardtii nuclear genes very well
(data not shown).
3.3. Deduced amino acid sequence and comparati6e analysis
The identified open reading frame (486 bp) en-codes a putative protein (GPXH) of 162 amino acids with a predicted molecular mass of 18.0 kDa. No potential signal peptide was found. The deduced amino acid sequence shows a high simi-larity (67 – 78% to the deduced amino acid
se-quence of the previously sequenced gpx
homologous genes of yeast (Fig. 2). It also exhibits significant homology to GPX-like proteins from several plants (between 60 and 65% similarity). Like these yeast and plant GPX-like proteins, C. reinhardtiiGPXH differs from the classic GPX1 in that it contains a Cys instead of the SeCys at the catalytic site. Apart from this difference, all GPX-like proteins contain the identical residues in the catalytic triad (Cys or SeCys, Gln, Trp) as well as identical amino acid regions in three protein loops, namely NVA…C (positions 70 – 75 in Fig. 2), L.FPCNQF…Q (positions 100 – 110), and WNF (positions 191 – 193) that are important for the catalytic activity in GPX1 [2].
The calculated similarities of GPXH from C.
reinhardtii with human GPX1 and GPX4 are
al-most equal (60 and 57%, respectively). C.
rein-hardtii GPXH (like the yeast and plant GPX-like proteins) resembles, however, the mammalian GPX4 more than GPX1 in that both GPXH and GPX4 lack the GSH binding residues identified in GPX1 and that a large loop (LMTDPKL-ITWSPVCR, positions 172 – 186 in Fig. 2 (human GPX1 sequence)) and a small helix (LNSL, posi-tions 119 – 122) are absent that have been shown to be important for subunit interaction in GPX1 [2].
3.4. Transcription start site and promoter of gpxh
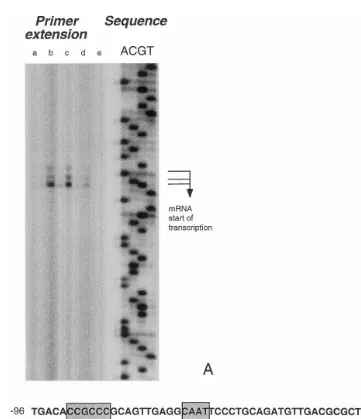
The transcription start site ofC.reinhardtii gpxh was determined in a primer extension experiment with several samples of total RNA extracted from C. reinhardtii (Fig. 3). Three subsequent otides (TGC) which are located 139 – 137 nucle-otides upstream of the coding sequence could be
identified as transcription start sites of gpxh
mRNA, the C residue being the most prominent. A smaller fraction of the mRNAs starts with G and even less mRNA is synthesized starting with T. The primer extension was carried out with several samples of total RNA known to contain
different relative amounts of the gpxh mRNA
(Northern blot analysis, see below). Since the
in-Fig. 1. Organization of gpxh gene and mRNA of C. reinhardtii. Numbers indicate the position of the site relative to the transcription start site. Boxed regions are the coding region (dark background) and introns (white background) within the coding region. Thick lines are exons, thin lines the chromosomal DNA upstream and downstream the transcribed region of thegpxh
Fig. 2. Alignment ofC.reinhardtiiGPXH amino acid sequence with the amino acid sequences of several other enzymes of the glutathione peroxidase family (Saccharomyces cere6isiae: unpublished, Acc. No. P38143 (swissprot); Citrus sinensis: [20];
Arabidopsis thalianaplastid-targeted GPX: unpublished, Acc. No. AJ000469 (swissprot);Nicotiana syl6estris: [19]; human GPX4: unpublished, Acc. No. X71973, mouse GPX5: [4]; human GPX1: [45]; nematode GPX: [10];Escherichia coliGPXH-like gene: [9]). The boxed sequences are: the catalytic triad (C or X, Q, and W; X indicates selenocysteine), glutathione binding residues in the human GPX1 (R and K) and signal peptides. Highlighted are amino acids homologous to C.reinhardtiiGPXH.
tensities of the detected bands were in agreement with the relative amounts of the transcript in the different samples (Fig. 4), false priming was ex-cluded. Primer extension experiments were also carried out with other primers and revealed the same transcription start sites with the same rela-tive signal intensities (data not shown).
Upstream from the transcription start site, a putative promoter region could be identified. This region contains the following elements that are common to eucaryotic RNA polymerase II pro-moters and that are located at the characteristic distances from the transcription start site in the
most prominent transcription start), two CAAT
boxes (−47 to −44 and −77 to −74) and a
GC-box (−93 to −88) (Fig. 3).
3.5. O6erexpression of gpxh in E. coli
In order to assign an enzymatic activity or a
physiological role to GPX, gpxh was
overex-pressed inE. coli BL21(DE3)(pLysS) to 10% of
the total cellular protein content (data not shown). Peroxidase activity was assayed in crude extracts using glutathione as electron donor and H2O2,
t-BOOH, or cumene hydroperoxide as electron
acceptors, basically following the protocol of the NADPH coupled assay described by Wendel [37]. Commercially available GPX was used as positive control. Unfortunately, no significant peroxidase activity could be measured. As an alternative ap-proach, we checked whether E. coli cells overex-pressing gpxh had an increased resistance towards oxidative stress conditions. The gpxh overexpress-ingE.coli cells did not reveal any increased oxida-tive stress resistance as compared to control cells (data not shown).
Fig. 3. Identification of thegpxhtranscription initiation site by primer extension and gpxhputative promoter region. Panel A: Primer extension ofgpxhmRNA. Primer extension and sequence were carried out using primer GPX5.4. Different lanes in the primer extension experiments are RNA samples taken from the same culture after exposure to 2 mM H2O2 at the following
Fig. 4. Northern blot analysis ofgpxhexpression after exposure to hydrogen peroxide. Total RNA was extracted from cultures ofC.reinhardtiicw15arg7exposed to 2 mM H2O2at the indicated time interval after addition. Of every sample 20mg of total RNA
were blotted and hybridized withgpxhcDNA as a probe.
3.6. Dynamics of the gpxh expression
In order to get a hint towards the physiological function of the C. reinhardtii gpxh gene, the
con-centration of the gpxh mRNA was monitored in
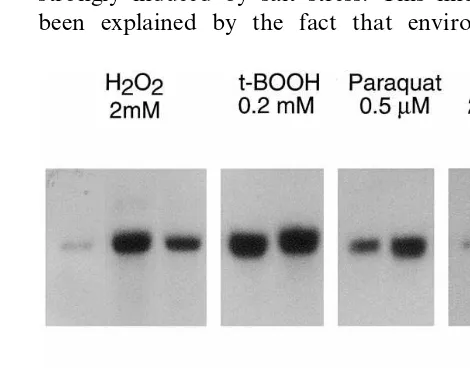
C. reinhardtii cells that were exposed to oxidative stress and to osmotic stress. Northern blot analysis withgpxhcDNA as a probe revealed that thegpxh mRNA content increased strongly after the
addi-tion of 2 mM H2O2 (Fig. 4) and reached a
maxi-mum level between 15 and 30 min after addition, being about eight times higher than that of the
unexposed cells. Subsequently, the gpxh mRNA
level decreased within one hour below the level of
unexposed cells. After 6 h, the gpxh mRNA level
of exposed cells equaled that of unexposed cells.
In order to test whether the gpxh mRNA levels
increased only upon oxidative stress or whether they increased due to a more general stress re-sponse, C. reinhardtii cells were treated with sub-lethal doses of compounds that are known to
cause oxidative stress (i.e. 0.2 mM t-BOOH, and
0.5 mM paraquat), or with 200 mM NaCl that
causes osmotic stress (Fig. 5). During exposure, cell numbers of the cultures did not significantly change, excluding an acute killing of the cells by the compounds. The agents excerting oxidative stress all lead to a strong accumulation of gpxh mRNA after 30 min and 3 h, whereas salt stress
did only result in a small change in gpxh mRNA
concentration within the time analyzed. This slight induction is in good agreement with the fact that also salt stress leads to the generation of perox-ides, to oxidative stress and increased levels of oxidative stress defensive enzymes [38,39].
4. Discussion
We report here the identification and characteri-zation of the single copy glutathione peroxidase homologous gene (gpxh) from C. reinhardtii.
The level of gpxh mRNA raised upon exposure of exponentially growing C. reinhardtii to chemi-cals known to cause oxidative stress, like hydrogen
peroxide, t-BOOH and paraquat [40]. Also salt
stress lead to small changes in the gpxh mRNA
level within 3 h. It has been shown previously that the gpx-related gene from Citrus sinensis is strongly induced by salt stress. This finding has been explained by the fact that environmental
Fig. 5. Northern blot analysis ofgpxhexpression upon expo-sure to various compounds. Total RNA was extracted from cultures ofC.reinhardtiicw15arg7exposed to 2 mM H2O2, 0.2
mMtert-butyl hydroperoxide, 0.5mM Paraquat, and 200 mM
NaCl, respectively, at the indicated time intervals after addi-tion. Of every sample 20 mg of total RNA were blotted and
stresses like drought and salt stress generate oxida-tive stress and indeed in many organisms lead to increased levels of mRNAs coding for enzymes involved in the defense against oxidative stress [5]. Our data that show a comparatively fast increase in the gpxh mRNA level upon exposure to H2O2 as compared to induction by NaCl are in good accordance with a model in which the response of plant GPX like proteins upon salt stress is medi-ated by reactive oxygen species [38].
The mRNA ofgpxhfromC.reinhardtiicontains
an open reading frame that codes for a putative GPX-like protein (GPXH) with a predicted molec-ular mass of 18.0 kDa which showed high homol-ogy to GPX-like proteins from yeast, plants and shared characteristic features with the mammalian SeCys dependent phospholipid hydroperoxide glu-tathione peroxidase (GPX4). GPXH as well as the yeast and plant GPX-like proteins contain a Cys residue at their putative active site in contrast to the well characterized mammalian GPX1 and GPX4 that contain a SeCys residue at their active site. The sulfur atom from Cys in principle is able to catalyze the same reactions as the selenium from SeCys, as a less powerful catalyst.
It is striking thatC.reinhardtiiGPXH, the plant and yeast GPX-like proteins as well as the mam-malian GPX4 lack the same amino acid stretches that are present in GPX1 (Fig. 2). In the case of the mammalian GPX4, the deletions disrupt the interfaces responsible for dimerization and te-tramerisation in GPX1 [2], which is in good accor-dance with the monomeric appearance of GPX4 and indicates that the GPX-like proteins isolated
from yeast, plants and GPXH from C. reinhardtii
probably occur in a monomeric form as well. Interestingly, the mammalian secretory Se-inde-pendent GPX5 do not lack the respective stretches and indeed occur as tetramers [41,42]. An addi-tional effect of the absence of these stretches of amino acids is that the active site is more freely accessible. This is consistent with the ability of GPX4 to attack more complex hydroperoxy lipids and with the weak activity of the GPX-like protein of C. sinensis towards phosphatidylcholine hy-droperoxide, whilst no activity towards hydrogen peroxide was measurable [17]. Similarly, weak GPX activities have been measured for the
Se-in-dependent GPX like proteins from pig andBrugia
pahangi [18,42]. However, Okamura et al. [42] found the GPX activity of GPX5 almost negligible
in the porcine epididymal fluid and proposed its physiological role to be rather binding to organic hydroperoxides than metabolizing them.
Our attempts to assign any GPX activity to
GPXH expressed in E. coli failed. Possibly, the
expression in E. coli does not lead to a functional protein, even though E. coli does contain a GPX-like protein itself [9]. Alternatively, GPXH activity might be restricted to more complex acceptor or donor substrates like different organic hydroper-oxides or large thiol compounds that are not metabolized by the other antioxidant enzymes present in C. reinhardtii, such as ascorbate peroxi-dase in the chloroplast, and catalase and a
te-trameric selenium-depending GPX in the
cytoplasm [7,43]. Furthermore, alternative roles for GPXH must be taken into consideration.
As an alternative function it has been speculated that some members of the GPX family like GPX4 might rather be involved in signal transduction than in the detoxification of hydroperoxides [44]. The flat hydrophobic surface surrounding the cat-alytic site of GPX4 would allow an interaction with both lipid layers and proteins. Thereby, oxi-dation equivalents from peroxides could be trans-ferred via the oxidized active site to protein thiol groups.
Although the northern blot analysis indicated a role of GPXH in the defense against oxidative stress, no GPX activity of GPXH overexpressed in
E. coli could be measured, nor did the
overex-pressed protein increase the oxidative stress resis-tance of E. colicells. Hence, the physiological role of this protein remains unclear. Even though for the GPX-like protein from C. sinensis, the induc-tion of mRNA upon oxidative stress, its
GPX4-like activity and the protection of E. coli
overexpressing the protein, clearly indicate that it is involved in the oxidative stress response, its exact role in the cell has neither been elucidated yet. A lot of effort has yet to be made to elucidate many unexplained observations concerning the role of the different Selenium independent GPX-like proteins that occur in organisms from all kingdoms. We believe that C. reinhardtii is a suit-able model organism to get more insight into the role of GPX-like proteins, because it contains both Selenium dependent GPX and Selenium indepen-dent GPX-like protein, because it is a
compara-tively simple haploid eucaryotic model and
Acknowledgements
We thank Dr Donald Weeks for generously
providing the cosmid library of C. reinhardtii
chromosomal DNA. We are also grateful to Car-ole Cossu, Nina Schweigert and Dr Sebastian Mendez-Alvarez for helpful suggestions through-out the course of the work.
References
[1] G.C. Mills, Glutathione peroxidase, an erythrocyte en-zyme which protects hemoglobin from oxidative break-down, J. Biol. Chem. 229 (1957) 189.
[2] F. Ursini, M. Maiorino, R. Brigelius-FlohZ, K.D. Au-mann, A. Roveri, D. Schomburg, L. FlohZ, Diversity of glutathione peroxidases, Methods Enzymol. 252 (1995) 38 – 53.
[3] L. Flohe, W.A. Gunzler, H.H. Schock, Glutathione per-oxidase: a selenoenzyme, FEBS Lett. 32 (1973) 132 – 134. [4] N. Ghyselinck, C. Jimenez, Y. Courty, J. Dufaure, An-drogen-dependent messenger RNA(s) related to secretory proteins in the mouse epididymis, J. Reprod. Fertil. 85 (1989) 631 – 639.
[5] Y. Eshdat, D. Holland, Z. Faltin, G. Benhayyim, Plant glutathione peroxidases, Physiol. Plant. 100 (1997) 234 – 240.
[6] L.T. Tran, Y. Inoue, A. Kimura, Oxidative stress re-sponse in yeast: purification and some properties of a membrane-bound glutathione peroxidase from
Hansenula mrakii, Biochim. Biophys. Acta 1164 (1993) 166 – 172.
[7] A. Yokota, S. Shigeoka, T. Onishi, S. Kitaoka, Selenium as inducer of glutathione peroxidase in low-CO2-grown
Chlamydomonas reinhardtii, Plant Physiol. 86 (1988) 649 – 651.
[8] E.L. Aho, L.P. Kelly, Identification of a glutathione peroxidase homolog inNeisseria meningitidis, DNA Seq. 6 (1995) 55 – 60.
[9] M.J. Friedrich, L.C. de Veaux, R.J. Kadner, Nucleotide sequence of the btuCED genes involved in vitamin B12 transport in Escherichia coliand homology with compo-nents of periplasmic-binding-protein-dependent transport systems, J. Bacteriol. 167 (1986) 928 – 934.
[10] E. Cookson, M.L. Blaxter, M.E. Selkirk, Identification of the major soluble cuticular glycoprotein of lymphatic filarial nematode parasites (gp29) as a secretory homolog of glutathione peroxidase, Proc. Natl. Acad. Sci. USA 89 (1992) 5837 – 5841.
[11] D.L. Williams, R.J. Pierce, E. Cookson, A. Capron, Molecular cloning and sequencing of glutathione peroxi-dase from Schistosoma mansoni, Mol. Biochem. Para-sitol. 52 (1992) 127 – 130.
[12] B. Gamain, J. Arnaud, A. Favier, D. Camus, D. Dive, C. Slomianny, Increase in glutathione peroxidase activity in malaria parasite after selenium supplementation, Free Radic. Biol. Med 21 (1996) 559 – 565.
[13] P. Roeckel Drevet, G. Gagne, D. de Labrouhe, J. Du-faure, P. Nicolas, J. Drevet, Molecular characterization, organ distribution and stress-mediated induction of two glutathione peroxidase-encoding mRNAs in sunflower (Helianthus annuus), Physiol. Plant. 103 (1998) 385 – 394. [14] P. Mullineaux, S. Karpinski, A. Jimenez, S. Cleary, C. Robinson, G. Creissen, Identification of cDNAs encod-ing plastid-targeted glutathione peroxidase, Plant J. 13 (1998) 375 – 379.
[15] N. Depege, J. Drevet, N. Boyer, Molecular cloning and characterization of tomato cDNAs encoding glutathione peroxidase-like proteins, Eur. J. Biochem. 253 (1998) 445 – 451.
[16] M. Maiorino, K.D. Aumann, R. Brigelius-Flohe, D. Doria, J. van den Heuvel, J. McCarthy, A. Roveri, F. Ursini, L. Flohe, Probing the presumed catalytic triad of selenium-containing peroxidases by mutational analysis of phospholipid hydroperoxide glutathione peroxidase (PHGPx), Biol. Chem. Hoppe Seyler 376 (1995) 651 – 660.
[17] T. Beeor-Tzahar, G. Benhayyim, D. Holland, Z. Faltin, Y. Eshdat, A stress associated citrus protein is a distinct plant phospholipid hydroperoxide glutathione peroxi-dase, FEBS Lett. 366 (1995) 151 – 155.
[18] L. Tang, K. Gounaris, C.M. Griffiths, M.E. Selkirk, Heterologous expression and enzymatic properties of a selenium-independent glutathione peroxidase from the parasitic nematode Brugia pahangi, J. Biol. Chem. 270 (1995) 18313 – 18318.
[19] M.C. Criqui, E. Jamet, Y. Parmentier, J. Marbach, A. Durr, J. Fleck, Isolation and characterization of a plant cDNA showing homology to animal glutathione peroxi-dases, Plant Mol. Biol. 18 (1992) 623 – 627.
[20] D. Holland, G. Ben-Hayyim, Z. Faltin, L. Camoin, A.D. Strosberg, Y. Eshdat, Molecular characterization of salt-stress-associated protein in citrus: protein and cDNA sequence homology to mammalian glutathione peroxi-dases, Plant Mol. Biol. 21 (1993) 923 – 927.
[21] D. Holland, Z. Faltin, A. Perl, G. Ben-Hayyim, Y. Eshdat, A novel plant glutathione peroxidase-like protein provides tolerance to oxygen radicals generated by paraquat in Escherichia coli, FEBS Lett. 337 (1994) 52 – 55.
[22] T. Moore, P.F. Sparling, Interruption of thegpxAgene increases the sensitivity of Neisseria meningitidis to paraquat, J. Bacteriol. 178 (1996) 4301 – 4305.
[23] P. Vernet, N. Rigaudiere, N. Ghyselinck, J. Dufaure, J. Drevet, In vitro expression of a mouse tissue specific glutathione-peroxidase-like protein lacking the selenocys-teine can protect stably transfected mammalian cells against oxidative damage, Biochem. Cell. Biol. 74 (1996) 125 – 131.
[24] N.J. Gumpel, J.D. Rochaix, S. Purton, Studies on ho-mologous recombination in the green alga Chlamy
-domonas reinhardtii, Curr. Genet. 26 (1994) 438 – 442. [25] J.D. Rochaix, Chlamydomonas reinhardtii as the
photo-synthetic yeast, Annu. Rev. Genet. 29 (1995) 209 – 230. [26] E.H. Harris, The Chlamydomonas Source Book. A
[27] J. Sambrook, E.F. Fritsch, T. Maniatis, Molecular Cloning, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, 1989.
[28] F.W. Studier, B.A. Moffatt, Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes, J. Mol. Biol. 189 (1986) 113 – 130. [29] A.H. Rosenberg, B.N. Lade, D.S. Chui, S.W. Lin, J.J.
Dunn, F.W. Studier, Vectors for selective expression of cloned DNAs by T7 RNA polymerase, Gene 56 (1987) 125 – 135.
[30] F.M. Ausubel, R. Brent, R.E. Kingston, D.D. Moore, J.G. Seidmann, J.A. Smith, K. Struhl, Current Protocols in Molecular Biology, Wiley, Massachusetts, 1994. [31] P. Chomczynski, N. Sacchi, Single-step method of RNA
isolation by acid guanidinium thiocyanate- phenol-chlo-roform extraction, Anal. Biochem. 162 (1987) 156 – 159. [32] H. Zhang, P.L. Herman, D.P. Weeks, Gene isolation
through genomic complenentation using an indexed li-brary of Chlamydomonas reinhardtii DNA, Plant Mol. Biol. 24 (1994) 663 – 672.
[33] M. Kozak, The scanning model for translation: an up-date, J. Cell Biol. 108 (1989) 229 – 241.
[34] S. Merchant, K. Hill, J.H. Kim, J. Thompson, D. Zaitlin, L. Bogorad, Isolation and characterization of a complementary DNA clone for an algal pre-apoplasto-cyanin, J. Biol. Chem. 265 (1990) 12372 – 12379. [35] S.M. Mount, A catalogue of splice junction sequences,
Nucleic Acids Res. 10 (1982) 459 – 472.
[36] Y. Nakamura, Chlamydomonas reinhardtii codon usage tabulated from genbank release 104. http://www. dna.affrc.go.jp/nakamura-bin/showcodon.cgi?species =Chlamydomonas+reihhardtii+[gbpln], 1998.
[37] A. Wendel, Glutathione peroxidase, Methods Enzymol. 77 (1981) 325 – 333.
[38] Y. Gueta-Dahan, Z. Yaniv, B.A. Zilinskas, G. Ben-Hayyim, Salt and oxidative stress: similar and specific responses and their relation to salt tolerance in Citrus, Planta 203 (1997) 460 – 469.
[39] M. Sugimoto, W. Sakamoto, Putative phospholipid hy-droperoxide glutathione peroxidase gene fromArabidop
-sis thaliana induced by oxidative stress, Genes Genet. Syst. 72 (1997) 311 – 316.
[40] B. Halliwell, J.M.C. Gutteridge, Free Radicals in Biology and Medicine, Clarendon Press, Oxford, 1989, pp. 1 – 543.
[41] A. Nieto, R. Gutierrez Sagal, M. Perez Martinez, Isola-tion and characterizaIsola-tion of a rabbit epididymal secre-tory glycoprotein that associates to the spermatozoon surface, Mol. Reprod. Dev. 46 (1997) 337 – 343. [42] N. Okamura, Y. Iwaki, S. Hiramoto, M. Tamba, S.
Bannai, Y. Sugita, P. Syntin, F. Dacheux, J. Dacheux, Molecular cloning and characterization of the epi-didymis-specific glutathione peroxidase-like protein secreted in the porcine epididymal fluid, Biochim. Bio-phys. Acta 1336 (1997) 99 – 109.
[43] T. Takeda, T. Ishikawa, S. Shigeoka, Metabolism of hydrogen peroxide by the scavenging system inChlamy
-domonas reinhardtii, Physiol. Plant. 99 (1997) 49 – 55. [44] F. Ursini, M. Maiorino, A. Roveri, Phospholipid
hy-droperoxide glutathione peroxidase (PHGPx): more than an antioxidant enzyme?, Biomed. Environ. Sci. 10 (1997) 327 – 332.
[45] K. Ishida, T. Morino, K. Takagi, Y. Sukenaga, Nucle-otide sequence of a human gene for glutathione peroxi-dase, Nucleic Acids Res. 15 (1987) 10051.
![Fig. 2. Alignment of Cunpublished, Acc. No. X71973, mouse GPX5: [4]; human GPX1: [45]; nematode GPX: [10];The boxed sequences are: the catalytic triad (C or X, Q, and W; X indicates selenocysteine), glutathione binding residues in thehuman GPX1 (R and K) a](https://thumb-ap.123doks.com/thumbv2/123dok/1035831.929144/6.612.59.506.16.518/alignment-cunpublished-sequences-catalytic-indicates-selenocysteine-glutathione-residues.webp)